



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (8): 24592. DOI: 10.17520/biods.2024592 cstr: 32101.14.biods.2024592
• Special Feature: Genetic Diversity and Conservation • Previous Articles Next Articles
Ruixiang Xue1,4, Xuerong Ma1,4, Jiongwen Wu1,4, Aijun Liu1,4, Xiquan Zhang1,4, Congliang Ji2, Yingshan Yin3, Weijian Zhu2, Qingbin Luo1,4,*( )
)
Received:2024-12-31
Accepted:2025-05-04
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: qbluo@scau.edu.cn.
Supported by:Ruixiang Xue, Xuerong Ma, Jiongwen Wu, Aijun Liu, Xiquan Zhang, Congliang Ji, Yingshan Yin, Weijian Zhu, Qingbin Luo. Genetic diversity and genetic structure of Zhongshan partridge duck populations[J]. Biodiv Sci, 2025, 33(8): 24592.
| 种类 Variety | 有效等位基因数 Ne | 观测杂合度 Ho | 期望杂合度 He | 多态信息含量 PIC | 核苷酸多样 π |
|---|---|---|---|---|---|
| 北京鸭 BJ | 3.9826 ± 2.3280 | 0.2633 ± 0.1229 | 0.3221 ± 0.1335 | 0.2645 ± 0.0918 | 0.0030 ± 0.0024 |
| 斑嘴鸭 BZ | 5.9716 ± 3.3542 | 0.1658 ± 0.1304 | 0.2389 ± 0.1392 | 0.2129 ± 0.1072 | 0.0034 ± 0.0028 |
| 东栏鸭 DL | 4.1970 ± 2.6863 | 0.3091 ± 0.1863 | 0.3201 ± 0.1394 | 0.2938 ± 0.1224 | 0.0028 ± 0.0024 |
| 奉化鸭 FH | 5.3629 ± 3.2577 | 0.1780 ± 0.1393 | 0.2661 ± 0.1437 | 0.2333 ± 0.1109 | 0.0028 ± 0.0023 |
| 枫叶鸭 FY | 4.1817 ± 2.4431 | 0.2789 ± 0.1824 | 0.3138 ± 0.1376 | 0.2865 ± 0.1201 | 0.0020 ± 0.0020 |
| 广西小麻鸭 GX | 4.2470 ± 2.6607 | 0.2902 ± 0.1591 | 0.3135 ± 0.1379 | 0.2708 ± 0.1035 | 0.0033 ± 0.0028 |
| 高邮鸭 GY | 3.6321 ± 2.1210 | 0.2597 ± 0.1479 | 0.3399 ± 0.1251 | 0.2908 ± 0.0896 | 0.0018 ± 0.0015 |
| 金定鸭 JD | 4.4242 ± 2.3207 | 0.1713 ± 0.1040 | 0.2865 ± 0.1272 | 0.2400 ± 0.0925 | 0.0001 ± 0.0001 |
| 靖西大麻鸭 JX | 3.6073 ± 1.8445 | 0.3134 ± 0.1512 | 0.3364 ± 0.1254 | 0.2881 ± 0.0915 | 0.0032 ± 0.0027 |
| 连城白鸭 LC | 4.6378 ± 2.3813 | 0.1771 ± 0.1074 | 0.2745 ± 0.1266 | 0.2317 ± 0.0926 | 0.0001 ± 0.0001 |
| 龙胜翠鸭 LS | 3.5011 ± 1.6571 | 0.3160 ± 0.1638 | 0.3407 ± 0.1234 | 0.2995 ± 0.0948 | 0.0033 ± 0.0026 |
| 莆田黑鸭 PT | 5.1533 ± 2.5464 | 0.1403 ± 0.1042 | 0.2495 ± 0.1241 | 0.2137 ± 0.0925 | 0.0001 ± 0.0001 |
| 融水香鸭 RS | 4.5350 ± 3.0489 | 0.2930 ± 0.1835 | 0.3084 ± 0.1440 | 0.2803 ± 0.1242 | 0.0029 ± 0.0026 |
| 山麻鸭 SM | 4.3646 ± 2.2142 | 0.1747 ± 0.0953 | 0.2863 ± 0.1241 | 0.2393 ± 0.0888 | 0.0001 ± 0.0001 |
| 绍兴鸭 SX | 3.7232 ± 2.1620 | 0.2186 ± 0.1196 | 0.3358 ± 0.1284 | 0.2750 ± 0.0873 | 0.0020 ± 0.0016 |
| 温氏麻鸭 WMA | 4.7271 ± 3.1733 | 0.3004 ± 0.1869 | 0.3009 ± 0.1460 | 0.2777 ± 0.1306 | 0.0034 ± 0.0029 |
| 文桥鸭 WQ | 4.4776 ± 3.0149 | 0.2976 ± 0.1829 | 0.3108 ± 0.1432 | 0.2826 ± 0.1231 | 0.0032 ± 0.0027 |
| 温氏白鸭 WSB | 4.3264 ± 2.9341 | 0.3048 ± 0.1847 | 0.3180 ± 0.1416 | 0.2897 ± 0.1266 | 0.0029 ± 0.0027 |
| 玉林麻鸭 YL | 4.6123 ± 3.1255 | 0.2916 ± 0.1832 | 0.3070 ± 0.1460 | 0.2781 ± 0.1248 | 0.0030 ± 0.0026 |
| 樱桃谷鸭 YTG | 4.0367 ± 2.3784 | 0.2378 ± 0.1587 | 0.3221 ± 0.1361 | 0.2799 ± 0.1034 | 0.0014 ± 0.0012 |
| 绿头野鸭 YY | 4.5105 ± 2.5598 | 0.2350 ± 0.1284 | 0.2922 ± 0.1364 | 0.2437 ± 0.0954 | 0.0037 ± 0.0027 |
| 中山麻鸭 ZS | 3.8656 ± 2.3322 | 0.3382 ± 0.1609 | 0.3313 ± 0.1329 | 0.2849 ± 0.1035 | 0.0032 ± 0.0029 |
Table 1 Genetic parameter table of population for 22 duck species/varieties
| 种类 Variety | 有效等位基因数 Ne | 观测杂合度 Ho | 期望杂合度 He | 多态信息含量 PIC | 核苷酸多样 π |
|---|---|---|---|---|---|
| 北京鸭 BJ | 3.9826 ± 2.3280 | 0.2633 ± 0.1229 | 0.3221 ± 0.1335 | 0.2645 ± 0.0918 | 0.0030 ± 0.0024 |
| 斑嘴鸭 BZ | 5.9716 ± 3.3542 | 0.1658 ± 0.1304 | 0.2389 ± 0.1392 | 0.2129 ± 0.1072 | 0.0034 ± 0.0028 |
| 东栏鸭 DL | 4.1970 ± 2.6863 | 0.3091 ± 0.1863 | 0.3201 ± 0.1394 | 0.2938 ± 0.1224 | 0.0028 ± 0.0024 |
| 奉化鸭 FH | 5.3629 ± 3.2577 | 0.1780 ± 0.1393 | 0.2661 ± 0.1437 | 0.2333 ± 0.1109 | 0.0028 ± 0.0023 |
| 枫叶鸭 FY | 4.1817 ± 2.4431 | 0.2789 ± 0.1824 | 0.3138 ± 0.1376 | 0.2865 ± 0.1201 | 0.0020 ± 0.0020 |
| 广西小麻鸭 GX | 4.2470 ± 2.6607 | 0.2902 ± 0.1591 | 0.3135 ± 0.1379 | 0.2708 ± 0.1035 | 0.0033 ± 0.0028 |
| 高邮鸭 GY | 3.6321 ± 2.1210 | 0.2597 ± 0.1479 | 0.3399 ± 0.1251 | 0.2908 ± 0.0896 | 0.0018 ± 0.0015 |
| 金定鸭 JD | 4.4242 ± 2.3207 | 0.1713 ± 0.1040 | 0.2865 ± 0.1272 | 0.2400 ± 0.0925 | 0.0001 ± 0.0001 |
| 靖西大麻鸭 JX | 3.6073 ± 1.8445 | 0.3134 ± 0.1512 | 0.3364 ± 0.1254 | 0.2881 ± 0.0915 | 0.0032 ± 0.0027 |
| 连城白鸭 LC | 4.6378 ± 2.3813 | 0.1771 ± 0.1074 | 0.2745 ± 0.1266 | 0.2317 ± 0.0926 | 0.0001 ± 0.0001 |
| 龙胜翠鸭 LS | 3.5011 ± 1.6571 | 0.3160 ± 0.1638 | 0.3407 ± 0.1234 | 0.2995 ± 0.0948 | 0.0033 ± 0.0026 |
| 莆田黑鸭 PT | 5.1533 ± 2.5464 | 0.1403 ± 0.1042 | 0.2495 ± 0.1241 | 0.2137 ± 0.0925 | 0.0001 ± 0.0001 |
| 融水香鸭 RS | 4.5350 ± 3.0489 | 0.2930 ± 0.1835 | 0.3084 ± 0.1440 | 0.2803 ± 0.1242 | 0.0029 ± 0.0026 |
| 山麻鸭 SM | 4.3646 ± 2.2142 | 0.1747 ± 0.0953 | 0.2863 ± 0.1241 | 0.2393 ± 0.0888 | 0.0001 ± 0.0001 |
| 绍兴鸭 SX | 3.7232 ± 2.1620 | 0.2186 ± 0.1196 | 0.3358 ± 0.1284 | 0.2750 ± 0.0873 | 0.0020 ± 0.0016 |
| 温氏麻鸭 WMA | 4.7271 ± 3.1733 | 0.3004 ± 0.1869 | 0.3009 ± 0.1460 | 0.2777 ± 0.1306 | 0.0034 ± 0.0029 |
| 文桥鸭 WQ | 4.4776 ± 3.0149 | 0.2976 ± 0.1829 | 0.3108 ± 0.1432 | 0.2826 ± 0.1231 | 0.0032 ± 0.0027 |
| 温氏白鸭 WSB | 4.3264 ± 2.9341 | 0.3048 ± 0.1847 | 0.3180 ± 0.1416 | 0.2897 ± 0.1266 | 0.0029 ± 0.0027 |
| 玉林麻鸭 YL | 4.6123 ± 3.1255 | 0.2916 ± 0.1832 | 0.3070 ± 0.1460 | 0.2781 ± 0.1248 | 0.0030 ± 0.0026 |
| 樱桃谷鸭 YTG | 4.0367 ± 2.3784 | 0.2378 ± 0.1587 | 0.3221 ± 0.1361 | 0.2799 ± 0.1034 | 0.0014 ± 0.0012 |
| 绿头野鸭 YY | 4.5105 ± 2.5598 | 0.2350 ± 0.1284 | 0.2922 ± 0.1364 | 0.2437 ± 0.0954 | 0.0037 ± 0.0027 |
| 中山麻鸭 ZS | 3.8656 ± 2.3322 | 0.3382 ± 0.1609 | 0.3313 ± 0.1329 | 0.2849 ± 0.1035 | 0.0032 ± 0.0029 |
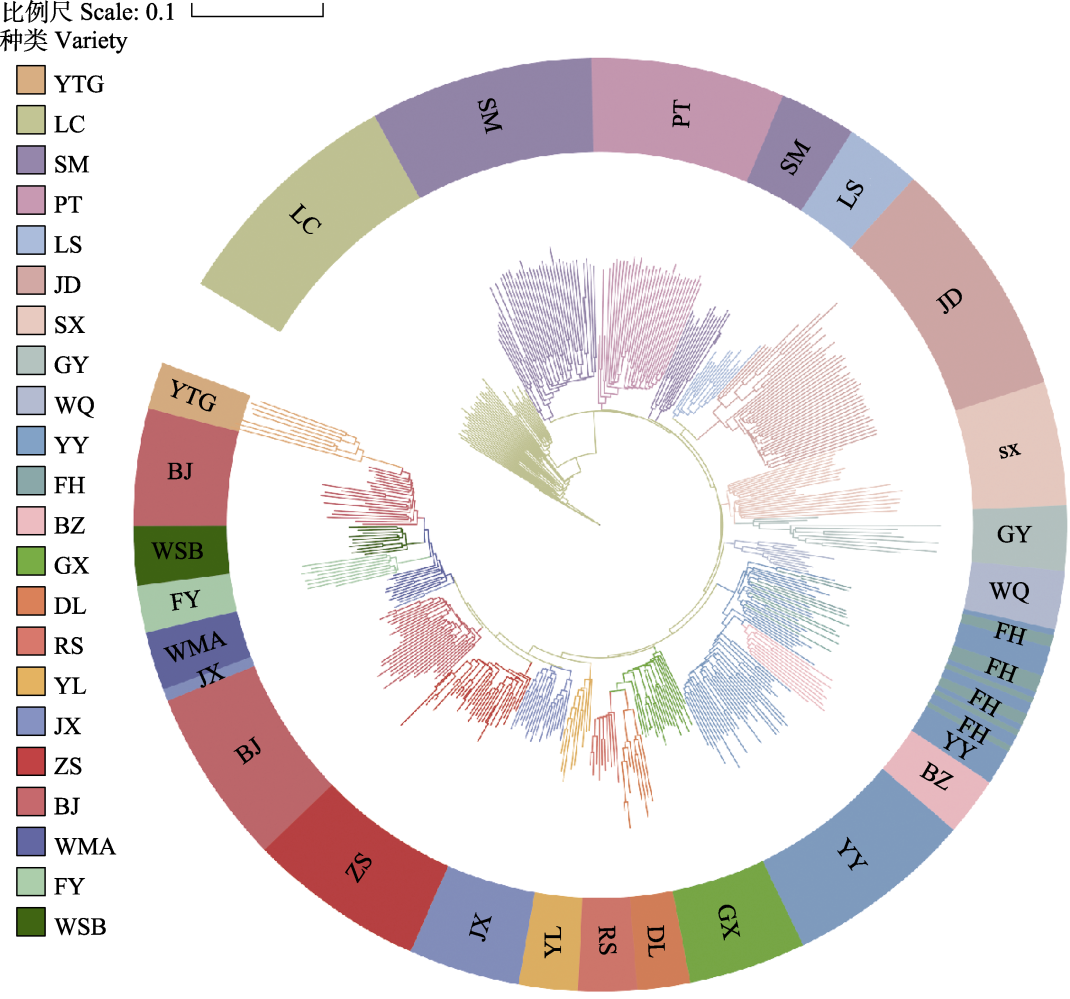
Fig. 1 Phylogenetic tree of 479 duck individuals. ZS, Zhongshan partridge duck; BJ, Beijing duck; LC, Liancheng white duck; WMA, Wenshi partridge duck; WSB, Wenshi white duck; GX, Guangxi small partridge duck; JX, Jingxi large partridge duck; YY, Mallard (Anas platyrhynchos); BZ, Spot-billed duck (Anas poecilorhyncha); FY, Maple leaf duck; YTG, Cherry valley duck; SX, Shaoxing duck; FH, Fenghua duck; GY, Gaoyou duck; SM, Mountain partridge duck; JD, Jinding duck; PT, Putian black duck; LS, Longsheng emerald duck; DL, Donglan duck; RS, Rongshui Xiang duck; WQ, Wenqiao duck; YL, Yulin partridge duck.
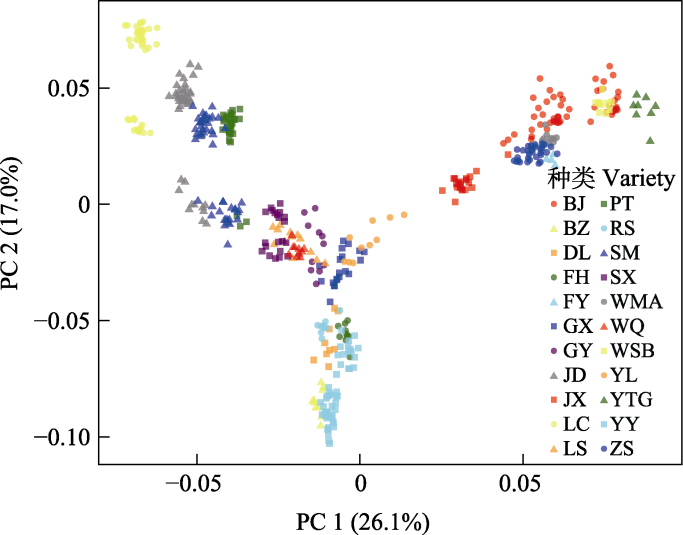
Fig. 2 Principal component analysis (PCA) plot of 479 duck individuals based on PC1 and PC2. Letters in the diagram represent abbreviations of duck breed names (ZS, Zhongshan partridge duck; BJ, Beijing duck; LC, Liancheng white duck; WMA, Wenshi partridge duck; WSB, Wenshi white duck; GX, Guangxi small partridge duck; JX, Jingxi large partridge duck; YY, Mallard (Anas platyrhynchos); BZ, Spot-billed duck (Anas poecilorhyncha); FY, Maple leaf duck; YTG, Cherry valley duck; SX, Shaoxing duck; FH, Fenghua duck; GY, Gaoyou duck; SM, Mountain partridge duck; JD, Jinding duck; PT, Putian black duck; LS, Longsheng emerald duck; DL, Donglan duck; RS, Rongshui Xiang duck; WQ, Wenqiao duck; YL, Yulin partridge duck).
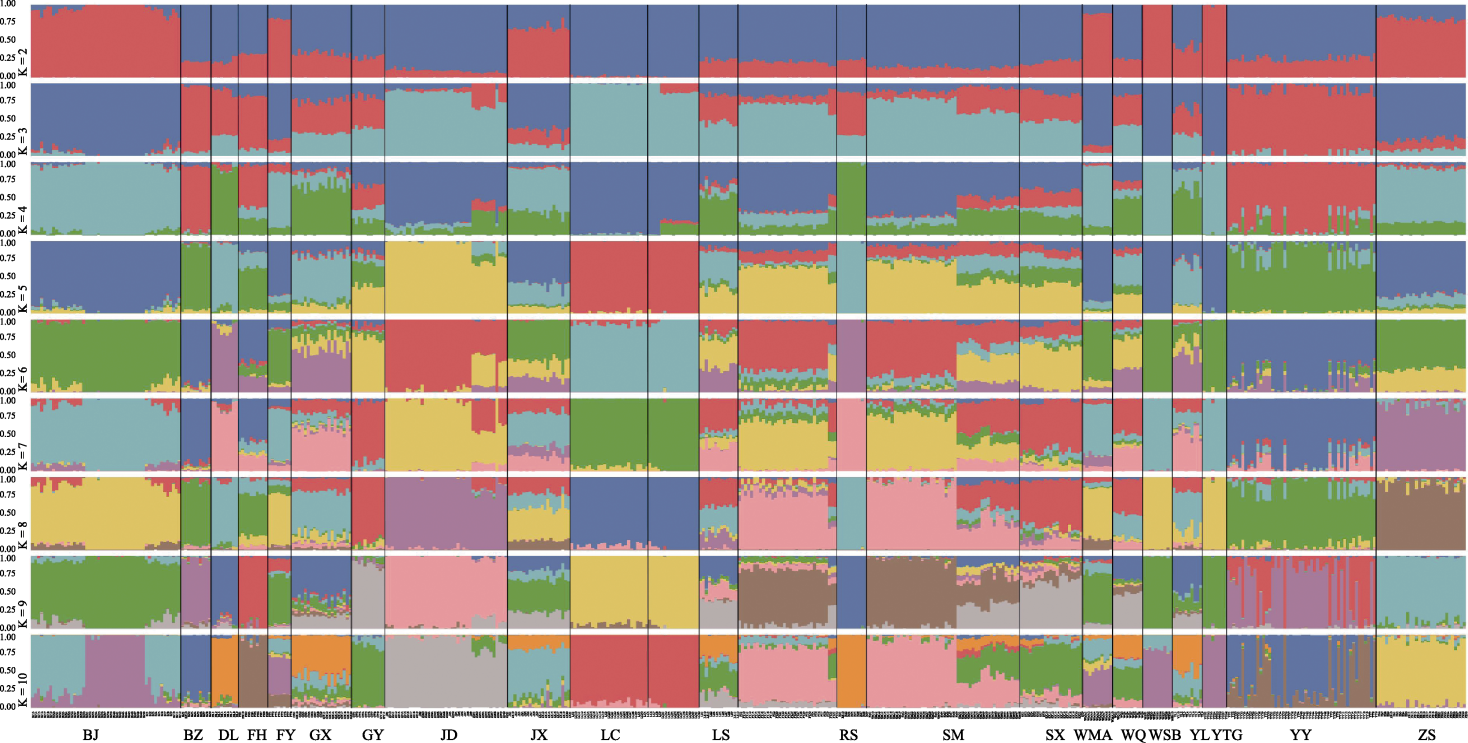
Fig. 3 Admixture analysis plot of 21 duck breeds from K = 2 to K = 10. ZS, Zhongshan partridge duck; BJ, Beijing duck; LC, Liancheng white duck; WMA, Wenshi partridge duck; WSB, Wenshi white duck; GX, Guangxi small partridge duck; JX, Jingxi large partridge duck; YY, Mallard (Anas platyrhynchos); BZ, Spot-billed duck (Anas poecilorhyncha); FY, Maple leaf duck; YTG, Cherry valley duck; SX, Shaoxing duck; FH, Fenghua duck; GY, Gaoyou duck; SM, Mountain partridge duck; JD, Jinding duck; PT, Putian black duck; LS, Longsheng emerald duck; DL, Donglan duck; RS, Rongshui Xiang duck; WQ, Wenqiao duck; YL, Yulin partridge duck.
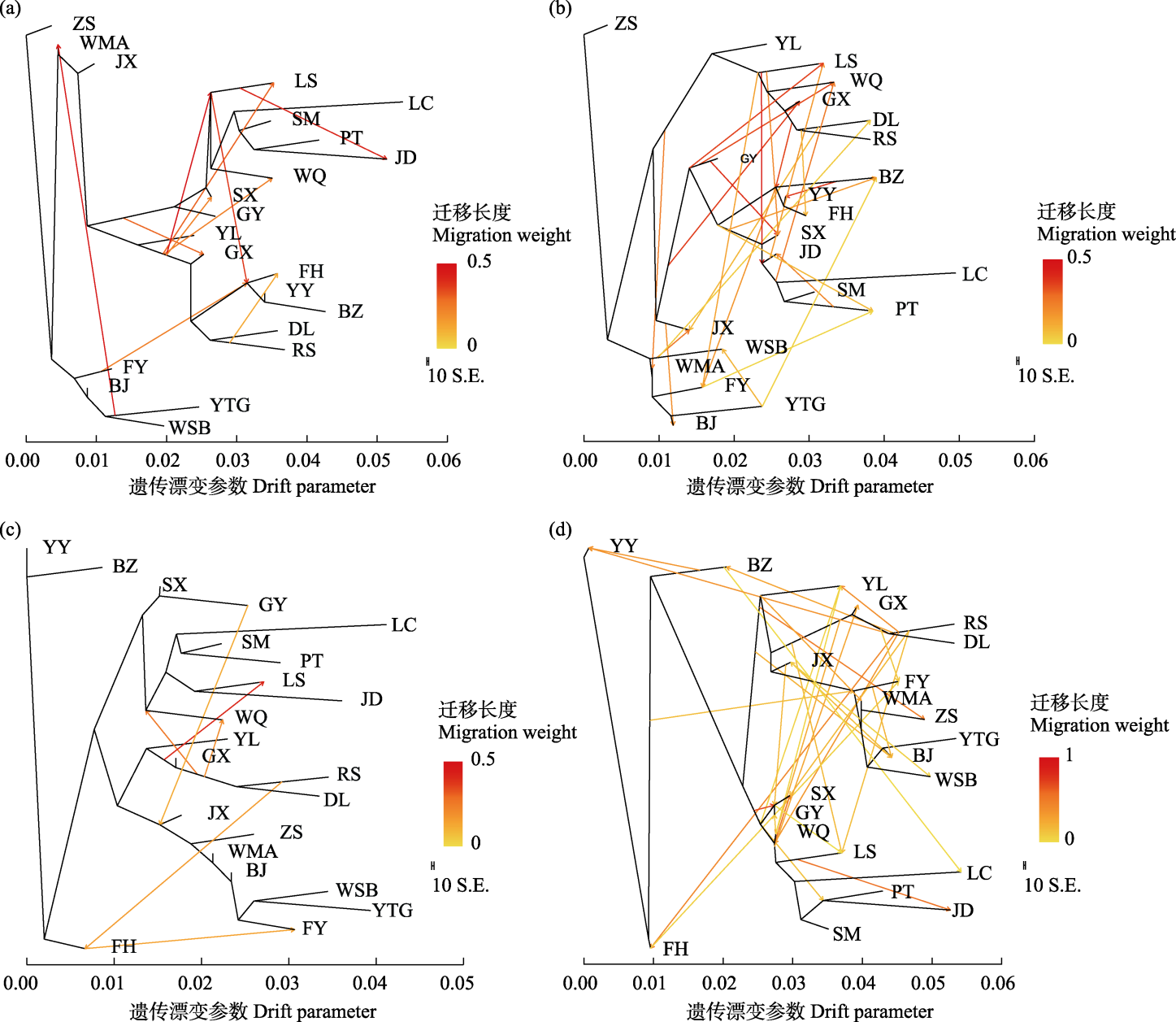
Fig. 4 TreeMix analysis plots of 22 duck breeds. (a) Maximum likelihood (ML) phylogenetic tree with Zhongshan partridge duck as the outgroup and 10 migration events. (b) ML phylogenetic tree with Zhongshan partridge duck as the outgroup and 24 migration events. (c) Maximum likelyhood phylogenetic tree with Mallard (Anas platyrhynchos) as the outgroup and 6 migration events. (d) ML phylogenetic tree with Mallard (Anas platyrhynchos) as the outgroup and 30 migration events. ZS, Zhongshan partridge duck; BJ, Beijing duck; LC, Liancheng white duck; WMA, Wenshi partridge duck; WSB, Wenshi white duck; GX, Guangxi small partridge duck; JX, Jingxi large partridge duck; YY, Mallard (Anas platyrhynchos); BZ, Spot-billed duck (Anas poecilorhyncha); FY, Maple leaf duck; YTG, Cherry valley duck; SX, Shaoxing duck; FH, Fenghua duck; GY, Gaoyou duck; SM, Mountain partridge duck; JD, Jinding duck; PT, Putian black duck; LS, Longsheng emerald duck; DL, Donglan duck; RS, Rongshui Xiang duck; WQ, Wenqiao duck; YL, Yulin partridge duck.
| [1] | Chen SF, Zhou YQ, Chen YR, Gu J (2018) Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34, i884-i890. |
| [2] | Chen ZJ, Wan YL, Zhong WJ, Chen SW, Zhou YH, Shi SJ, Jiang L, Ji PC, Yang ZH, Mao ZX, Mou FS (2021) Development and application of soybean in del markers based on whole-genome resequencing datasets. Journal of Plant Genetic Resources, 22, 815-833. (in Chinese with English abstract) |
|
[陈正杰, 宛永璐, 钟文娟, 陈四维, 周永航, 石盛佳, 蒋理, 戢沛城, 杨泽湖, 毛正轩, 牟方生 (2021) 基于大豆基因组重测序的InDel标记开发及应用. 植物遗传资源学报, 22, 815-833.]
DOI |
|
| [3] |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Group GPA (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156-2158.
DOI PMID |
| [4] |
Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, Li ST, Larkin DM, Kim H, Frantz LAF, Caccamo M, Ahn H, Aken BL, Anselmo A, Anthon C, Auvil L, Badaoui B, Beattie CW, Bendixen C, Berman D, Blecha F, Blomberg J, Bolund L, Bosse M, Botti S, Zhan BJ, Bystrom M, Capitanu B, Carvalho-Silva D, Chardon P, Chen C, Cheng R, Choi SH, Chow W, Clark RC, Clee C, Crooijmans RPMA, Dawson HD, Dehais P, De Sapio F, Dibbits B, Drou N, Du ZQ, Eversole K, Fadista J, Fairley S, Faraut T, Faulkner GJ, Fowler KE, Fredholm M, Fritz E, Gilbert JGR, Giuffra E, Gorodkin J, Griffin DK, Harrow JL, Hayward A, Howe K, Hu ZL, Humphray SJ, Hunt T, Hornshøj H, Jeon JT, Jern P, Jones M, Jurka J, Kanamori H, Kapetanovic R, Kim J, Kim JH, Kim KW, Kim TH, Larson G, Lee K, Lee KT, Leggett R, Lewin HA, Li YR, Liu WS, Loveland JE, Lu Y, Lunney JK, Ma J, Madsen O, Mann K, Matthews L, McLaren S, Morozumi T, Murtaugh MP, Narayan J, Nguyen DT, Ni PX, Oh SJ, Onteru S, Panitz F, Park EW, Park HS, Pascal G, Paudel Y, Perez-Enciso M, Ramirez-Gonzalez R, Reecy JM, Rodriguez-Zas S, Rohrer GA, Rund L, Sang YM, Schachtschneider K, Schraiber JG, Schwartz J, Scobie L, Scott C, Searle S, Servin B, Southey BR, Sperber G, Stadler P, Sweedler JV, Tafer H, Thomsen B, Wali R, Wang J, Wang J, White S, Xu X, Yerle M, Zhang GJ, Zhang JG, Zhang J, Zhao SH, Rogers J, Churcher C, Schook LB (2012) Analyses of pig genomes provide insight into porcine demography and evolution. Nature, 491, 393-398.
DOI |
| [5] |
Li HF, Xu WJ, Zhu WQ, Xue SM, Chen KW (2008) Genetic diversity and evolution of Fujian domestic duck breeds. Biodiversity Science, 16, 607-612. (in Chinese with English abstract)
DOI |
|
[李慧芳, 徐文娟, 朱文奇, 薛素美, 陈宽维 (2008) 福建省家鸭的遗传多样性及其与野鸭的亲缘关系. 生物多样性, 16, 607-612.]
DOI |
|
| [6] |
Lin RY, Li JQ, Yang Y, Yang YH, Chen JM, Zhao FL, Xiao TF (2022) Genome-wide population structure analysis and genetic diversity detection of four Chinese indigenous duck breeds from Fujian Province. Animals, 12, 2302.
DOI URL |
| [7] |
Lin Y, Huang M, Li XJ, Zhang XM, Huang YM, Tian YB, Wu ZP (2023) Uncovering genome-wide copy number variations in 8 duck breeds using whole genome resequencing data. Acta Veterinaria et Zootechnica Sinica, 54, 3700-3709. (in Chinese with English abstract)
DOI |
|
[林燕, 黄敏, 李秀金, 张续勐, 黄运茂, 田允波, 伍仲平 (2023) 利用全基因组重测序数据检测8个鸭品种基因组拷贝数变异. 畜牧兽医学报, 54, 3700-3709.]
DOI |
|
| [8] |
Liu Z, Sun H, Lai W, Hu M, Zhang Y, Bai C, Liu J, Ren H, Li F, Yan S (2022) Genome-wide re-sequencing reveals population structure and genetic diversity of Bohai black cattle. Animal Genetics, 53, 133-136.
DOI URL |
| [9] | Luo W (2021) Genetic Diversity of Genome of Local Chicken Breeds in China and Analysis of Selection Imprint of Cockfighting and Antlers’ Crown Populations. PhD dissertation, South China Agricultural University, Guangzhou. (in Chinese with English abstract) |
| [罗威 (2021) 中国家鸡地方品种基因组遗传多样性及斗鸡、鹿角冠群体的选择印记分析. 博士毕业论文, 华南农业大学, 广州.] | |
| [10] |
Luo W, Luo CL, Wang M, Guo LJ, Chen XL, Li ZH, Zheng M, Folaniyi BS, Luo W, Shu DM, Song LL, Fang MX, Zhang XQ, Qu H, Nie QH (2020) Genome diversity of Chinese indigenous chicken and the selective signatures in Chinese gamecock chicken. Scientific Reports, 10, 14532.
DOI PMID |
| [11] | Mishra S, Sarkar U, Taraphder S, Datta S, Swain D, Saikhom R, Sasmita P, Menalsh L (2017) Multivariate statistical data analysis—Principal component analysis (PCA). International Journal of Livestock Research, 7, 60-78. |
| [12] | Pickrell JK, Pritchard JK (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics, 8, e1002967. |
| [13] |
Tubelyte V, Švažas S, Sruoga A, Butkauskas D, Paulauskas A, Baublys V, Viksne J, Grishanov G, Kozulin A (2011) Genetic diversity of tufted ducks (Aythya fuligula, Anatidae) in Eastern Europe. Open Life Sciences, 6, 1044-1053.
DOI URL |
| [14] | Wang R, Sun JL, Han H, Huang YF, Chen T, Yang MM, Wei Q, Wan HF, Liao YY (2021) Whole-genome resequencing reveals genetic characteristics of different duck breeds from the Guangxi region in China. G3: Genes, Genomes, Genetics, 11, jkab054. |
| [15] | Wang TM, Guo QX, Bai H, Chang GB, Chen GH (2021) Analysis of population structure of different duck genetic resources based on whole-genome resequencing. Chinese Journal of Animal Science, 57(11), 78-81. (in Chinese with English abstract) |
| [王统苗, 郭其新, 白皓, 常国斌, 陈国宏 (2021) 基于全基因组重测序对不同鸭遗传资源进行群体结构分析. 中国畜牧杂志, 57(11), 78-81.] | |
| [16] |
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution, 38, 1358-1370.
DOI PMID |
| [17] |
Wu PX, Wang K, Yang Q, Zhou J, Chen DJ, Liu YH, Ma JD, Tang QZ, Jin L, Xiao WH, Lou PE, Jiang AN, Jiang YZ, Zhu L, Li MZ, Li XW, Tang GQ (2019) Whole-genome re-sequencing association study for direct genetic effects and social genetic effects of six growth traits in large white pigs. Scientific Reports, 9, 9667.
DOI PMID |
| [18] | Zhang L (2019) Genome Sequencing and Research of Bama Mini-Pig. PhD dissertation, Guangxi University, Nanning. (in Chinese with English abstract) |
| [张笠 (2019) 广西巴马小型猪全基因组测序及分析研究. 博士学位论文, 广西大学, 南宁.] | |
| [19] | Zhang Y (2014) Genetic Diversity and Population Structure Analysis of Some Local Duck Breeds in China. PhD dissertation, Yangzhou University, Yangzhou, Jiangsu. (in Chinese with English abstract) |
| [张扬 (2014) 我国部分地方鸭品种遗传多样性与群体结构分析. 博士学位论文, 扬州大学, 江苏扬州.] | |
| [20] |
Zhang YH, Jia HX, Wang ZB, Sun P, Cao DM, Hu JJ (2019) Genetic diversity and population structure of Populus yunnanensis. Biodiversity Science, 27, 355-365. (in Chinese with English abstract)
DOI URL |
|
[张亚红, 贾会霞, 王志彬, 孙佩, 曹德美, 胡建军 (2019) 滇杨种群遗传多样性与遗传结构. 生物多样性, 27, 355-365.]
DOI |
|
| [21] | Zhang ZB (2018) Study on Selection Signal and Domestication Time of Domestic Ducks by Genome-wide Resequencing. PhD dissertation, China Agricultural University, Beijing. (in Chinese with English abstract) |
| [张泽宾 (2018) 利用全基因组重测序研究家鸭驯化的选择信号及驯化时间. 博士学位论文, 中国农业大学, 北京.] |
| [1] | Ping Fan, Zhixin Wen, Gang Song. The effect of climatic factors and anthropogenic activities on different genetic diversity indicators of amphibians and mammals [J]. Biodiv Sci, 2025, 33(8): 25022-. |
| [2] | Ping Fan, Huan Wang, Zhixin Wen, Gang Song, Fuming Lei. Impact of climatic factors on the genetic diversity-species area relationship of birds [J]. Biodiv Sci, 2025, 33(8): 25072-. |
| [3] | Qilong Yu, Minhui Hao, Huaijiang He, Chunyu Zhang, Xiuhai Zhao. Relationships of biodiversity and productivity change with forest succession in Changbai Mountains: Insights from species, traits, and phylogeny [J]. Biodiv Sci, 2025, 33(8): 25060-. |
| [4] | Zhicheng Zhou, Tianling Cao, Ruyao Liu, Qiqi Ding, Ke Ma, Liping Yang, Chuanjiang Zhou, Guoxing Nie, Yongtao Tang. Genetic diversity and structure of Pseudorasbora parva population in the Yellow River basin based on the mitochondrial COI gene [J]. Biodiv Sci, 2025, 33(8): 24501-. |
| [5] | Ruxiao Wang, Boyang Shi, Da Pan, Hongying Sun. Biodiversity patterns and conservation gaps of the endemic freshwater crab genus Sinopotamon in China [J]. Biodiv Sci, 2025, 33(8): 25123-. |
| [6] | Huixia Li, Yu Li, Xin Ning, Xiaochen Li, Tianrui Wang, Yigang Song, Xiling Dai, Sisi Zheng, Xin Zhong. Genetic diversity and genetic structure of Gymnospermium kiangnanense based on chloroplast genome [J]. Biodiv Sci, 2025, 33(8): 25149-. |
| [7] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [8] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [9] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [10] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [11] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [12] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [13] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [14] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [15] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()