



生物多样性 ›› 2025, Vol. 33 ›› Issue (2): 24192. DOI: 10.17520/biods.2024192 cstr: 32101.14.biods.2024192
曹东1,2,3( ), 李焕龙1,2,3(
), 李焕龙1,2,3( ), 彭扬1,2,3(
), 彭扬1,2,3( ), 魏存争1,2,3,*(
), 魏存争1,2,3,*( )(
)( )
)
收稿日期:2024-05-20
接受日期:2024-08-01
出版日期:2025-02-20
发布日期:2024-11-05
通讯作者:
*E-mail: weicunzheng@ibcas.ac.cn
基金资助:
Cao Dong1,2,3( ), Li Huanlong1,2,3(
), Li Huanlong1,2,3( ), Peng Yang1,2,3(
), Peng Yang1,2,3( ), Wei Cunzheng1,2,3,*(
), Wei Cunzheng1,2,3,*( )(
)( )
)
Received:2024-05-20
Accepted:2024-08-01
Online:2025-02-20
Published:2024-11-05
Contact:
*E-mail: weicunzheng@ibcas.ac.cn
Supported by:摘要:
物种的基因组大小反映了基因组扩张和收缩机制之间的动态平衡, 其受自然选择和遗传漂变等多重因素影响。关于基因组大小与性状关系的研究成果众多, 但尚缺乏系统性的梳理和总结。本文总结了植物基因组大小与41个性状之间的60种不同关联, 并尝试将这些性状划分为9个尺度, 包括分子、细胞、组织、器官、个体、种群、群落、生态系统和生物地理尺度。为解释基因组大小与性状之间的关系, 本文在现有假说的基础上, 提出了“基因组大小分区假说”。该假说认为, 植物基因组大小的分布范围内可能存在两个平衡点和一个最优点, 而环境条件如养分和水分的变化, 可能会改变这些平衡点和最优点的位置, 从而驱动植物基因组大小向新的最优点方向演化, 形成新的平衡。最后, 本文建议将基因组大小作为核心功能性状纳入生态模型, 以提升对群落和生态系统应对全球环境挑战(如氮沉降和气候变化)的理解。
曹东, 李焕龙, 彭扬, 魏存争 (2025) 植物基因组大小与性状关系的研究进展. 生物多样性, 33, 24192. DOI: 10.17520/biods.2024192.
Cao Dong, Li Huanlong, Peng Yang, Wei Cunzheng (2025) Progresses in the study of the relationship between plant genome size and traits. Biodiversity Science, 33, 24192. DOI: 10.17520/biods.2024192.
| 尺度 Scale | 性状 Traits | 关系 Correlation | 参考文献 References |
|---|---|---|---|
| 分子 Molecule | 转座子数 Transposables | + | Wang et al, |
| ^ | Lynch et al, | ||
| GC含量 GC content | + | Musto et al, | |
| ^ | Veselý et al, | ||
| 染色体数量 Chromosome number | + | Wang et al, | |
| 细胞 Cell | 细胞周期长度 Cell cycle duration | + | Bennett, |
| 细胞大小 Cell size | + | Renzaglia et al, | |
| 花粉大小 Pollen size | + | Knight et al, | |
| 细胞密度 Cell density | ‒ | Théroux-Rancourt et al, | |
| 细胞分裂速率 Cell division rate | ‒ | Bennett & Smith, | |
| 细胞核体积 Nucleus size | + | Van’t Hof & Sparrow, | |
| 精子移动能力 Sperm mobility | ‒ | Renzaglia et al, | |
| 组织 Tissue | 叶片组织弹性 Leaf tissue elasticity | ‒ | Castro-Jimenez et al, |
| 导管直径 Xylem vessel diameter | + | Maherali et al, | |
| 器官 Organ | 种子重量 Seed mass | + | Maranon & Grubb, |
| ns | Leishman, | ||
| 种子数量 Seed number | ‒ | Suda et al, | |
| 叶片长度 Leaf length | + | Chung et al, | |
| ‒ | Caceres et al, | ||
| 叶片宽度 Leaf width | ‒ | Zonneveld & Duncan, | |
| + | Chung et al, | ||
| 叶片含氮量 Leaf nitrogen content | + | Kang et al, | |
| 叶片含磷量 Leaf phosphorus content | ns | Kang et al, | |
| 比叶面积 Specific leaf area | + | Kang et al, | |
| 开花时间 Flowering period | ‒ | Veselý et al, | |
| 个体 Individual | 生长速率 Growth rate | + | Grime et al, |
| ‒ | Biradar et al, | ||
| 最短代际时间 Minimum generation time | + | Chooi, | |
| ‒ | Labani & Elkingtion, | ||
| ns | Cavallini et al, | ||
| 最大植株高度 Maximum plant height | ‒ | 邵晨等, | |
| 最大光合速率 Maximum photosynthetic rate | ‒ | Beaulieu et al, | |
| 代谢速率 Metabolic rate | ‒ | Szarski, | |
| 种群 Population | 种群大小 Population size | ‒ | Charlesworth & Barton, |
| 入侵能力 Invasiveness | ‒ | Chen et al, | |
| ns | Gallagher et al, | ||
| 物种分化速率 Diversification rate | ‒ | Kraaijeveld, | |
| 灭绝速率 Risk of extinction | + | Vinogradov, | |
| 群落 Community | 群落竞争性策略 Community competitive strategy | ^ | Šmarda et al, |
| 物种丰富度 Species richness | ^ | Peng et al, | |
| 群落生产力 Community productivity | ^ | Šmarda et al, | |
| 群落霜冻耐受性 Community frost resistance | + | Macgillivray & Grime, | |
| 生态系统 Ecosystem | 磷可利用性 Phosphorus availability | + | Šmarda et al, |
| 氮可利用性 Nitrogen availability | + | Kang et al, | |
| CO2 浓度 CO2 concentration | + | Jasieński & Bazzaz, | |
| 重金属离子浓度 Heavy metal concentration | ‒ | Vidic et al, | |
| 温度 Temperature | + | Turpeinen et al, | |
| ‒ | Campbell et al, | ||
| ^ | Knight & Ackerly, | ||
| ns | Kalendar et al, | ||
| 降雨量 Precipitation | + | Price et al, | |
| ‒ | Bottini et al, | ||
| ns | Sims & Price, | ||
| 生物地理 Biogeography | 纬度 Latitude | + | Miksche, |
| ‒ | Bennett et al, | ||
| ns | Teoh & Rees, | ||
| 海拔高度 Altitude | + | Bennett, | |
| ‒ | Mangelsdorf & Cameron, | ||
| ^ | Rayburn, | ||
| ns | Palomino, |
表1 基因组大小与不同性状之间的关系(+代表正相关, ‒代表负相关, ^代表非线性, ns代表不相关)
Table 1 Relationships between genome size and various traits (+ indicates positive correlation, ‒ indicates negative correlation, ^ indicates non-linear relationship, ns indicates no significant correlation)
| 尺度 Scale | 性状 Traits | 关系 Correlation | 参考文献 References |
|---|---|---|---|
| 分子 Molecule | 转座子数 Transposables | + | Wang et al, |
| ^ | Lynch et al, | ||
| GC含量 GC content | + | Musto et al, | |
| ^ | Veselý et al, | ||
| 染色体数量 Chromosome number | + | Wang et al, | |
| 细胞 Cell | 细胞周期长度 Cell cycle duration | + | Bennett, |
| 细胞大小 Cell size | + | Renzaglia et al, | |
| 花粉大小 Pollen size | + | Knight et al, | |
| 细胞密度 Cell density | ‒ | Théroux-Rancourt et al, | |
| 细胞分裂速率 Cell division rate | ‒ | Bennett & Smith, | |
| 细胞核体积 Nucleus size | + | Van’t Hof & Sparrow, | |
| 精子移动能力 Sperm mobility | ‒ | Renzaglia et al, | |
| 组织 Tissue | 叶片组织弹性 Leaf tissue elasticity | ‒ | Castro-Jimenez et al, |
| 导管直径 Xylem vessel diameter | + | Maherali et al, | |
| 器官 Organ | 种子重量 Seed mass | + | Maranon & Grubb, |
| ns | Leishman, | ||
| 种子数量 Seed number | ‒ | Suda et al, | |
| 叶片长度 Leaf length | + | Chung et al, | |
| ‒ | Caceres et al, | ||
| 叶片宽度 Leaf width | ‒ | Zonneveld & Duncan, | |
| + | Chung et al, | ||
| 叶片含氮量 Leaf nitrogen content | + | Kang et al, | |
| 叶片含磷量 Leaf phosphorus content | ns | Kang et al, | |
| 比叶面积 Specific leaf area | + | Kang et al, | |
| 开花时间 Flowering period | ‒ | Veselý et al, | |
| 个体 Individual | 生长速率 Growth rate | + | Grime et al, |
| ‒ | Biradar et al, | ||
| 最短代际时间 Minimum generation time | + | Chooi, | |
| ‒ | Labani & Elkingtion, | ||
| ns | Cavallini et al, | ||
| 最大植株高度 Maximum plant height | ‒ | 邵晨等, | |
| 最大光合速率 Maximum photosynthetic rate | ‒ | Beaulieu et al, | |
| 代谢速率 Metabolic rate | ‒ | Szarski, | |
| 种群 Population | 种群大小 Population size | ‒ | Charlesworth & Barton, |
| 入侵能力 Invasiveness | ‒ | Chen et al, | |
| ns | Gallagher et al, | ||
| 物种分化速率 Diversification rate | ‒ | Kraaijeveld, | |
| 灭绝速率 Risk of extinction | + | Vinogradov, | |
| 群落 Community | 群落竞争性策略 Community competitive strategy | ^ | Šmarda et al, |
| 物种丰富度 Species richness | ^ | Peng et al, | |
| 群落生产力 Community productivity | ^ | Šmarda et al, | |
| 群落霜冻耐受性 Community frost resistance | + | Macgillivray & Grime, | |
| 生态系统 Ecosystem | 磷可利用性 Phosphorus availability | + | Šmarda et al, |
| 氮可利用性 Nitrogen availability | + | Kang et al, | |
| CO2 浓度 CO2 concentration | + | Jasieński & Bazzaz, | |
| 重金属离子浓度 Heavy metal concentration | ‒ | Vidic et al, | |
| 温度 Temperature | + | Turpeinen et al, | |
| ‒ | Campbell et al, | ||
| ^ | Knight & Ackerly, | ||
| ns | Kalendar et al, | ||
| 降雨量 Precipitation | + | Price et al, | |
| ‒ | Bottini et al, | ||
| ns | Sims & Price, | ||
| 生物地理 Biogeography | 纬度 Latitude | + | Miksche, |
| ‒ | Bennett et al, | ||
| ns | Teoh & Rees, | ||
| 海拔高度 Altitude | + | Bennett, | |
| ‒ | Mangelsdorf & Cameron, | ||
| ^ | Rayburn, | ||
| ns | Palomino, |
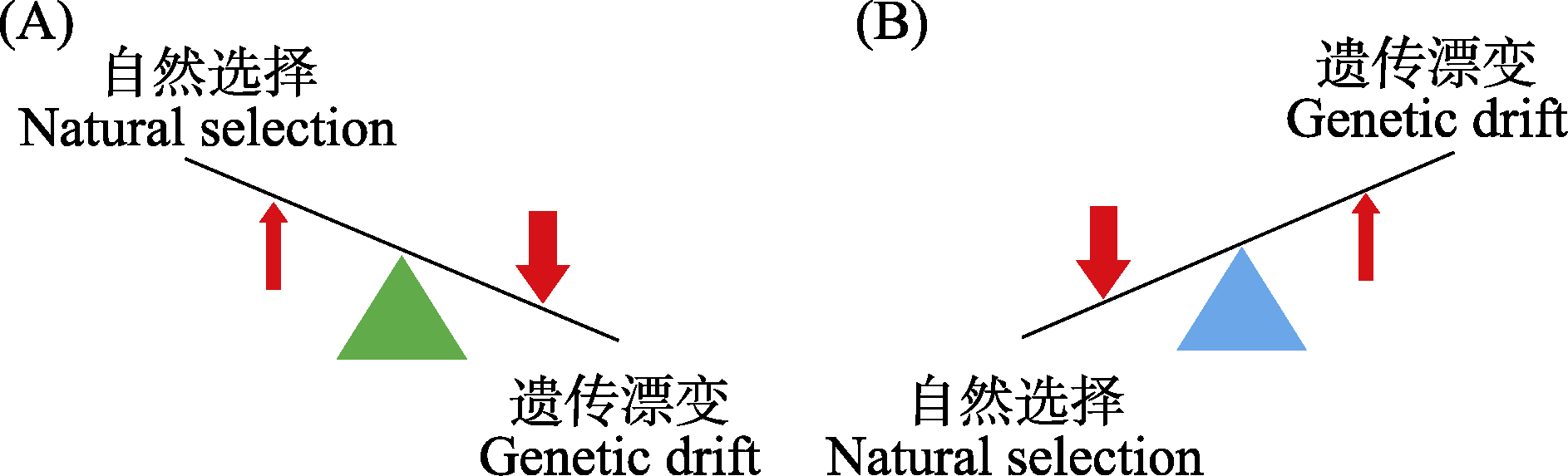
图2 自然选择与遗传漂变两种力量对基因组大小演化的影响, 改编自Blommaert (2020)。(A)遗传漂变在基因组大小的演化中扮演的角色更重要; (B)自然选择在基因组大小的演化中扮演的角色更重要。
Fig. 2 Two scenarios illustrating the relative roles of natural selection and genetic drift in genome size evolution, adapted from Blommaert (2020). (A) Genetic drift plays a more significant role in genome size evolution; (B) Natural selection plays a more sigmificant role in genome size evolution.
| 优势 Advantages | 劣势 Disadvantages | |
|---|---|---|
| 大基因组 Larger GS | 对霜冻的耐受性更强 Greater resistance to frost (Macgillivray & Grime, 提供自然选择的功能空间更多 More function space for natural selection (Mei et al, 后代的遗传多样性更高 Greater genetic diversity in offspring (石米娟等, | 对养分的需求量更大 Greater demand for nutrients (Guignard et al, 修复DNA的能量需求更多 More energy needed to repair DNA (Hidalgo et al, 生长速度更慢 Slower growth rates (Hessen et al, |
| 小基因组 Smaller GS | 对养分的需求量更少 Lower demand for nutrients (Guignard et al, 修复DNA的能量需求更多 Less energy needed to repair DNA (Hidalgo et al, 生长速度更快 Faster growth rates (Hessen et al, | 对霜冻的耐受性更弱 Weaker resistance to frost (Macgillivray & Grime, 提供自然选择的功能空间更少 Less function space for natural selection (Mei et al, 后代的遗传多样性更低 Lower genetic diversity in offspring (石米娟等, |
表2 大基因组和小基因组各自的优劣势(GS指基因组大小)
Table 2 Some advantages and disadvantages of larger genome and smaller genome (GS means genome size)
| 优势 Advantages | 劣势 Disadvantages | |
|---|---|---|
| 大基因组 Larger GS | 对霜冻的耐受性更强 Greater resistance to frost (Macgillivray & Grime, 提供自然选择的功能空间更多 More function space for natural selection (Mei et al, 后代的遗传多样性更高 Greater genetic diversity in offspring (石米娟等, | 对养分的需求量更大 Greater demand for nutrients (Guignard et al, 修复DNA的能量需求更多 More energy needed to repair DNA (Hidalgo et al, 生长速度更慢 Slower growth rates (Hessen et al, |
| 小基因组 Smaller GS | 对养分的需求量更少 Lower demand for nutrients (Guignard et al, 修复DNA的能量需求更多 Less energy needed to repair DNA (Hidalgo et al, 生长速度更快 Faster growth rates (Hessen et al, | 对霜冻的耐受性更弱 Weaker resistance to frost (Macgillivray & Grime, 提供自然选择的功能空间更少 Less function space for natural selection (Mei et al, 后代的遗传多样性更低 Lower genetic diversity in offspring (石米娟等, |
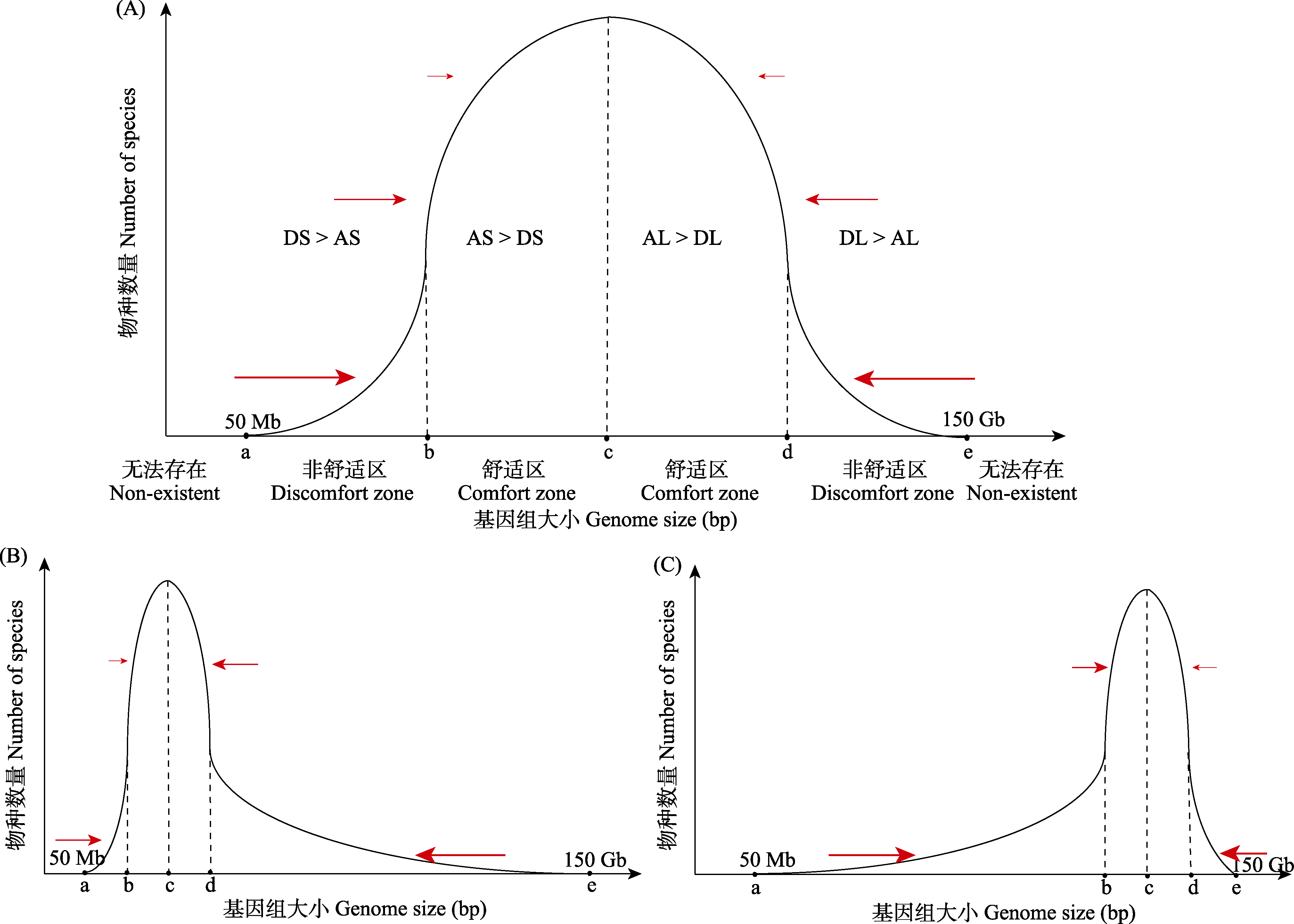
图3 三种可能存在的基因组大小分布模式。红色箭头代表环境因素施加的选择压力, 箭头越大代表选择压力越强。DS: 基因组小的劣势; AS: 基因组小的优势; AL: 基因组大的优势; DL: 基因组大的劣势。
Fig. 3 Three potential distribution patterns of genome size in angiosperms. The red arrows indicate the selection pressure exerted by environmental factors, with larger arrows representing stronger pressures. DS, Disadvantages of smaller genome size; AS, Advantages of smaller genome size; AL, Advantages of larger genome size; DL, Disadvantages of larger genome size.
| [1] |
Atkinson RRL, Mockford EJ, Bennett C, Christin PA, Spriggs EL, Freckleton RP, Thompson K, Rees M, Osborne CP (2016) C4 photosynthesis boosts growth by altering physiology, allocation and size. Nature Plants, 2, 16038.
DOI PMID |
| [2] | Baetcke KP, Sparrow AH, Nauman CH, Schwemmer SS (1967) The relationship of DNA content to nuclear and chromosome volumes and to radiosensitivity (LD50). Proceedings of the National Academy of Sciences, USA, 58, 533-540. |
| [3] | Baranyi M, Greilhuber J (1999) Genome size in Allium: In quest of reproducible data. Annals of Botany, 83, 687-695. |
| [4] |
Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA (2008) Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist, 179, 975-986.
DOI PMID |
| [5] |
Beaulieu JM, Moles AT, Leitch IJ, Bennett MD, Dickie JB, Knight CA (2007) Correlated evolution of genome size and seed mass. New Phytologist, 173, 422-437.
PMID |
| [6] | Bennett MD (1972) Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences, 181, 109-135. |
| [7] | Bennett MD (1976) DNA amount, latitude, and crop plant distribution. Environmental and Experimental Botany, 16, 93-108. |
| [8] | Bennett MD (1987) Variation in genomic form in plants and its ecological implications. New Phytologist, 106, 177-200. |
| [9] |
Bennett MD (1996) The nucleotype, the natural karyotype and the ancestral genome. Symposia of the Society for Experimental Biology, 50, 45-52.
PMID |
| [10] | Bennett MD, Bhandol P, Leitch IJ (2000) Nuclear DNA amounts in angiosperms and their modern uses— 807 new estimates. Annals of Botany, 86, 859-909. |
| [11] |
Bennett MD, Leitch IJ (2005) Plant genome size research: A field in focus. Annals of Botany, 95, 1-6.
PMID |
| [12] | Bennett MD, Smith JB (1972) The effects of polyploidy on meiotic duration and pollen development in cereal anthers. Proceedings of the Royal Society B: Biological Sciences, 181, 81-107. |
| [13] | Bennett MD, Smith JB, Lewis Smith RI (1982) DNA amounts of angiosperms from the Antarctic and South Georgia. Environmental and Experimental Botany, 22, 307-318. |
| [14] |
Biradar DP, Bullock DG, Rayburn AL (1994) Nuclear DNA amount, growth, and yield parameters in maize. Theoretical and Applied Genetics, 88, 557-560.
DOI PMID |
| [15] | Blommaert J (2020) Genome size evolution:Towards new model systems for old questions. Proceedings of the Royal Society B: Biological Sciences, 287, 20201441. |
| [16] | Bottini MCJ, Greizerstein EJ, Aulicino MB, Poggio L (2001) Relationships among genome size, environmental conditions and geographical distribution in natural populations of NW Patagonian species of Berberis L. (Berberidaceae). Annals of Botany, 86, 565-573. |
| [17] | Bretagnolle F, Thompson JD (1996) An experimental study of ecological differences in winter growth between sympatric diploid and autotetraploid Dactylis glomerata. Journal of Ecology, 84, 343-351. |
| [18] | Bureš P, Elliott TL, Veselý P, Šmarda P, Forest F, Leitch IJ, Nic Lughadha E, Soto Gomez M, Pironon S, Brown MJM, Šmerda J, Zedek F (2024) The global distribution of angiosperm genome size is shaped by climate. New Phytologist, 242, 744-759. |
| [19] | Caceres ME, De Pace C, Scarascia Mugnozza GT, Kotsonis P, Ceccarelli M, Cionini PG (1998) Genome size variations within Dasypyrum villosum: Correlations with chromosomal traits, environmental factors and plant phenotypic characteristics and behaviour in reproduction. Theoretical and Applied Genetics, 96, 559-567. |
| [20] | Campbell BD, Caradus JR, Hunt CL (1999) Temperature responses and nuclear DNA amounts of seven white clover populations which differ in early spring growth rates. New Zealand Journal of Agricultural Research, 42, 9-17. |
| [21] | Castro-Jimenez Y, Newton RJ, Price HJ, Halliwell RS (1989) Drought stress responses of Microseris species differing in nuclear DNA content. American Journal of Botany, 76, 789-795. |
| [22] |
Cavalier-Smith T (1978) Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. Journal of Cell Science, 34, 247-278.
PMID |
| [23] |
Cavalier-Smith T (1982) Skeletal DNA and the evolution of genome size. Annual Review of Biophysics and Bioengineering, 11, 273-302.
PMID |
| [24] | Cavalier-Smith T (1985) DNA replication and the evolution of genome size. In: The Evolution of Genome Size (ed. Cavalier-Smith T), pp. 211-251. John Wiley & Sons, Chichester. |
| [25] |
Cavalier-Smith T (2005) Economy, speed and size matter: Evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany, 95, 147-175.
PMID |
| [26] |
Cavalier-Smith T, Beaton MJ (1999) The skeletal function of non-genic nuclear DNA: New evidence from ancient cell chimaeras. Genetica, 106, 3-13.
PMID |
| [27] | Cavallini A, Natali L, Cionini G, Gennai D (1993) Nuclear DNA variability within Pisum sativum (Leguminosae): Nucleotypic effects on plant growth. Heredity, 70, 561-565. |
| [28] |
Ceccarelli M, Falistocco E, Cionini PG (1992) Variation in genome size and organization within hexaploid Festuca arundinaceae. Theoretical and Applied Genetics, 83, 273-278.
DOI PMID |
| [29] | Cerbah M, Coulaud J, Brown SC, Siljak-Yakovlev S (1999) Evolutionary DNA variation in the genus Hypochaeris. Heredity, 82, 261-266. |
| [30] | Charlesworth B, Barton N (2004) Genome size: Does bigger mean worse?. Current Biology, 14, R233-R235. |
| [31] | Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature, 371, 215-220. |
| [32] | Chen GQ, Guo SL, Yin LP (2010) Applying DNA C-values to evaluate invasiveness of angiosperms: Validity and limitation. Biological Invasions, 12, 1335-1348. |
| [33] | Chen JJ, Wang Y (2009) Recent progress in plant genome size evolution. Hereditas, 31, 464-470. (in Chinese with English abstract) |
| [ 陈建军, 王瑛 (2009) 植物基因组大小进化的研究进展. 遗传, 31, 464-470.] | |
| [34] |
Chipman AD, Khaner O, Haas A, Tchernov E (2001) The evolution of genome size: What can be learned from anuran development? Journal of Experimental Zoology, 291, 365-374.
PMID |
| [35] |
Chooi WY (1971) Variation in nuclear DNA content in the genus Vicia. Genetics, 68, 195-211.
DOI PMID |
| [36] | Chung J, Lee JH, Arumuganathan K, Graef GL, Specht JE (1998) Relationships between nuclear DNA content and seed and leaf size in soybean. Theoretical and Applied Genetics, 96, 1064-1068. |
| [37] | Collas P (1998) Cytoplasmic control of nuclear assembly. Reproduction Fertility and Development, 10, 581-592. |
| [38] |
Creber HMC, Davies MS, Francis D, Walker HD (1994) Variation in DNA C value in natural populations of Dactylis glomerata. New Phytologist, 128, 555-561.
DOI PMID |
| [39] |
D’Ario M, Tavares R, Schiessl K, Desvoyes B, Gutierrez C, Howard M, Sablowski R (2021) Cell size controlled in plants using DNA content as an internal scale. Science, 372, 1176-1181.
DOI PMID |
| [40] |
De Baerdemaeker NJF, Hias N, Van den Bulcke J, Keulemans W, Steppe K (2018) The effect of polyploidization on tree hydraulic functioning. American Journal of Botany, 105, 161-171.
DOI PMID |
| [41] | Dodsworth S, Leitch AR, Leitch IJ (2015) Genome size diversity in angiosperms and its influence on gene space. Current Opinion in Genetics & Development, 35, 73-78. |
| [42] |
Dufresne F, Jeffery N (2011) A guided tour of large genome size in animals: What we know and where we are heading. Chromosome Research, 19, 925-938.
DOI PMID |
| [43] |
Faizullah L, Morton JA, Hersch-Green EI, Walczyk AM, Leitch AR, Leitch IJ (2021) Exploring environmental selection on genome size in angiosperms. Trends in Plant Science, 26, 1039-1049.
DOI PMID |
| [44] | Fernández P, Amice R, Bruy D, Christenhusz MJM, Leitch IJ, Leitch AL, Pokorny L, Hidalgo O, Pellicer J (2024) A 160 Gbp fork fern genome shatters size record for eukaryotes. iScience, 27, 109889. |
| [45] |
Fleischmann A, Michael TP, Rivadavia F, Sousa A, Wang WQ, Temsch EM, Greilhuber J, Müller KF, Heubl G (2014) Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Annals of Botany, 114, 1651-1663.
DOI PMID |
| [46] | Gallagher RV, Leishman MR, Miller JT, Hui C, Richardson DM, Suda J, Trávníček P (2011) Invasiveness in introduced Australian acacias: The role of species traits and genome size. Diversity and Distributions, 17, 884-897. |
| [47] | Ghannoum O, Evans JR, von Caemmerer S (2010) Nitrogen and water use efficiency of C4 plants. In: C4 Photosynthesis and Related CO2 Concentrating Mechanisms (eds Raghavendra A, Sage R), pp. 129-146. Springer, Dordrecht, Netherlands. |
| [48] |
Godelle B, Cartier D, Marie D, Brown SC, Siljak-Yakovlev S (1993) Heterochromatin study demonstrating the non-linearity of fluorometry useful for calculating genomic base composition. Cytometry, 14, 618-626.
PMID |
| [49] |
Gregory TR (2001) Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews, 76, 65-101.
PMID |
| [50] |
Gregory TR (2002) A bird’s-eye view of the C-value enigma: Genome size, cell size, and metabolic rate in the class Aves. Evolution, 56, 121-130.
PMID |
| [51] |
Gregory TR (2005) Synergy between sequence and size in large-scale genomics. Nature Reviews Genetics, 6, 699-708.
DOI PMID |
| [52] |
Gregory TR, Hebert PD (1999) The modulation of DNA content: Proximate causes and ultimate consequences. Genome Research, 9, 317-324.
PMID |
| [53] |
Greilhuber J, Doležel J, Lysák MA, Bennett MD (2005) The origin, evolution and proposed stabilization of the terms ‘Genome Size’ and ‘C-Value’ to describe nuclear DNA contents. Annals of Botany, 95, 255-260.
PMID |
| [54] | Grime JP, Shacklock JML, Band SR (1985) Nuclear DNA contents, shoot phenology and species co-existence in a limestone grassland community. New Phytologist, 100, 435-445. |
| [55] |
Grotkopp E, Rejmánek M, Sanderson MJ, Rost TL (2004) Evolution of genome size in pines (Pinus) and its life-history correlates: Supertree analyses. Evolution, 58, 1705-1729.
PMID |
| [56] | Guignard MS, Crawley MJ, Kovalenko D, Nichols RA, Trimmer M, Leitch AR, Leitch IJ (2019) Interactions between plant genome size, nutrients and herbivory by rabbits, molluscs and insects on a temperate grassland. Proceedings of the Royal Society B: Biological Sciences, 286, 20182619. |
| [57] |
Guignard MS, Nichols RA, Knell RJ, Macdonald A, Romila CA, Trimmer M, Leitch IJ, Leitch AR (2016) Genome size and ploidy influence angiosperm species’ biomass under nitrogen and phosphorus limitation. New Phytologist, 210, 1195-1206.
DOI PMID |
| [58] | Hao GY, Lucero ME, Sanderson SC, Zacharias EH, Holbrook NM (2013) Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae). New Phytologist, 197, 970-978. |
| [59] | Henry TA, Bainard JD, Newmaster SG (2015) Genome size evolution in Ontario ferns (Polypodiidae): Evolutionary correlations with cell size, spore size, and habitat type and an absence of genome downsizing. Genome, 57, 555-566. |
| [60] | Hessen DO, Jeyasingh PD, Neiman M, Weider LJ (2010) Genome streamlining and the elemental costs of growth. Trends in Ecology & Evolution, 25, 75-80. |
| [61] |
Hidalgo O, Pellicer J, Christenhusz M, Schneider H, Leitch AR, Leitch IJ (2017) Is there an upper limit to genome size? Trends in Plant Science, 22, 567-573.
DOI PMID |
| [62] |
Horner HA, Macgregor HC (1983) C value and cell volume: Their significance in the evolution and development of amphibians. Journal of Cell Science, 63, 135-146.
PMID |
| [63] | Jasieński M, Bazzaz FA (1995) Genome size and high CO2. Nature, 376, 559-560. |
| [64] | Jockusch EL (1997) An evolutionary correlate of genome size change in plethodontid salamanders. Proceedings of the Royal Society B: Biological Sciences, 264, 597-604. |
| [65] | Jones RN, Brown LM (1976) Chromosome evolution and DNA variation in Crepis. Heredity, 36, 91-104. |
| [66] |
Jones RN, Viegas W, Houben A (2008) A century of B chromosomes in plants: So what? Annals of Botany, 101, 767-775.
DOI PMID |
| [67] | Joyner KL, Wang XR, Johnston JS, Price HJ, Williams CG (2001) DNA content for Asian pines parallels New World relatives. Canadian Journal of Botany, 79, 192-196. |
| [68] | Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences, USA, 97, 6603-6607. |
| [69] |
Kang M, Tao JJ, Wang J, Ren C, Qi QW, Xiang QY, Huang HW (2014) Adaptive and nonadaptive genome size evolution in Karst endemic flora of China. New Phytologist, 202, 1371-1381.
DOI PMID |
| [70] |
Kang M, Wang J, Huang HW (2015) Nitrogen limitation as a driver of genome size evolution in a group of karst plants. Scientific Reports, 5, 11636.
DOI PMID |
| [71] | Knight CA, Ackerly DD (2002) Variation in nuclear DNA content across environmental gradients: A quantile regression analysis. Ecology Letters, 5, 66-76. |
| [72] |
Knight CA, Beaulieu JM (2008) Genome size scaling through phenotype space. Annals of Botany, 101, 759-766.
DOI PMID |
| [73] | Knight CA, Clancy RB, Götzenberger L, Dann L, Beaulieu JM (2010) On the relationship between pollen size and genome size. Journal of Botany, 2010, 612017. |
| [74] |
Knight CA, Molinari NA, Petrov DA (2005) The large genome constraint hypothesis: Evolution, ecology and phenotype. Annals of Botany, 95, 177-190.
DOI PMID |
| [75] | Kozłowski J, Konarzewski M, Gawelczyk AT (2003) Cell size as a link between noncoding DNA and metabolic rate scaling. Proceedings of the National Academy of Sciences, USA, 100, 14080-14085. |
| [76] |
Kraaijeveld K (2010) Genome size and species diversification. Evolutionary Biology, 37, 227-233.
PMID |
| [77] | Kubešová M, Moravcová L, Suda J, Jarošik V, Pyšek P (2010) Naturalized plants have smaller genomes than their non-invading relatives: A flow cytometric analysis of the Czech alien flora. Preslia, 82, 81-96. |
| [78] | Labani RM, Elkington TT (1987) Nuclear DNA variation in the genus Allium L. (Liliaceae) Heredity, 59, 119-128. |
| [79] | Laurie DA, Bennett MD (1985) Nuclear DNA content in the genera Zea and Sorghum. Intergeneric, interspecific and intraspecific variation. Heredity, 55, 307-313. |
| [80] |
Lavergne S, Muenke NJ, Molofsky J (2010) Genome size reduction can trigger rapid phenotypic evolution in invasive plants. Annals of Botany, 105, 109-116.
DOI PMID |
| [81] | Leishman MR (1999) How well do plant traits correlate with establishment ability? Evidence from a study of 16 calcareous grassland species. New Phytologist, 141, 487-496. |
| [82] |
Leitch AR, Leitch IJ (2008) Genomic plasticity and the diversity of polyploid plants. Science, 320, 481-483.
DOI PMID |
| [83] | Leitch IJ, Leitch AR (2013) Genome size diversity and evolution in land plants. In: Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes (eds Leitch IJ, Greilhuber J, Doležel J, Wende JF), pp. 307-322. Springer, Wien, Germany. |
| [84] | Lertzman-Lepofsky G, Mooers AØ, Greenberg DA (2019) Ecological constraints associated with genome size across salamander lineages. Proceedings of the Royal Society B: Biological Sciences, 286, 20191780. |
| [85] | Levin DA, Funderburg SW (1979) Genome size in angiosperms: Temperate versus tropical species. American Naturalist, 114, 784-795. |
| [86] |
Lomax BH, Hilton J, Bateman RM, Upchurch GR, Lake JA, Leitch IJ, Cromwell A, Knight CA (2014) Reconstructing relative genome size of vascular plants through geological time. New Phytologist, 201, 636-644.
DOI PMID |
| [87] |
Lynch M, Koskella B, Schaack S (2006) Mutation pressure and the evolution of organelle genomic architecture. Science, 311, 1727-1730.
DOI PMID |
| [88] | Lysák MA, Rostková A, Dixon JM, Rossi G, Doležel J (2000) Limited genome size variation in Sesleria albicans. Annals of Botany, 86, 399-403. |
| [89] | Macgillivray CW, Grime JP (1995) Genome size predicts frost resistance in British herbaceous plants: Implications for rates of vegetation response to global warming. Functional Ecology, 9, 320-325. |
| [90] |
Maherali H, Walden AE, Husband BC (2009) Genome duplication and the evolution of physiological responses to water stress. New Phytologist, 184, 721-731.
DOI PMID |
| [91] | Mangelsdorf PC, Cameron JW (1942) Western Guatemala: A secondary center of origin of cultivated maize varieties. Botanical Museum Leaflets, Harvard University, 10, 217-255. |
| [92] | Maranon T, Grubb PJ (1993) Physiological basis and ecological significance of the seed size and relative growth rate relationship in Mediterranean annuals. Functional Ecology, 7, 591-599. |
| [93] | Maynard JS, Szathmary E (1995) The major transitions in evolution. In: Encyclopedia of Evolution (ed. Pagel M), pp. 17-22. . Oxford University Press, Oxford. |
| [94] |
Mei WB, Stetter MG, Gates DJ, Stitzer MC, Ross-Ibarra J (2018) Adaptation in plant genomes: Bigger is different. American Journal of Botany, 105, 16-19.
DOI PMID |
| [95] | Meyerson LA, Pyšek P, Lučanová M, Wigginton S, Tran CT, Cronin JT (2020) Plant genome size influences stress tolerance of invasive and native plants via plasticity. Ecosphere, 11, e03145. |
| [96] | Miksche JP (1967) Variation in DNA content of several gymnosperms. Canadian Journal of Genetics and Cytology, 9, 717-722. |
| [97] | Miksche JP (1971) Intraspecific variation of DNA per cell between Picea sitchensis (Bong.) Carr. provenances. Chromosoma, 32, 343-352. |
| [98] | Minelli S, Moscariello P, Ceccarelli M, Cionini PG (1996) Nucleotype and phenotype in Vicia faba. Heredity, 76, 524-530. |
| [99] |
Musto H, Naya H, Zavala A, Romero H, Alvarez-Valín F, Bernardi G (2006) Genomic GC level, optimal growth temperature, and genome size in Prokaryotes. Biochemical and Biophysical Research Communications, 347, 1-3.
PMID |
| [100] |
Novák P, Guignard MS, Neumann P, Kelly LJ, Mlinarec J, Koblížková A, Dodsworth S, Kovařík A, Pellicer J, Wang WC, Macas J, Leitch IJ, Leitch AR (2020) Repeat-sequence turnover shifts fundamentally in species with large genomes. Nature Plants, 6, 1325-1329.
DOI PMID |
| [101] | Orgel LE, Crick FH (1980) Selfish DNA: The ultimate parasite. Nature, 284, 604-607. |
| [102] | Pagel M, Johnstone RA (1992) Variation across species in the size of the nuclear genome supports the junk-DNA explanation for the C-value paradox. Proceedings of the Royal Society B: Biological Sciences, 249, 119-124. |
| [103] | Palomino G (1993) DNA content for seven diploid species and 569 populations of Echeandia (Lilliaceae). American Journal of Botany, 80, 77. |
| [104] | Palomino G, Sousa SM (2000) Variation of nuclear DNA content in the biflorus species of Lonchocarpus(Leguminosae). Annals of Botany, 85, 69-76. |
| [105] |
Pandit MK, White SM, Pocock MJO (2014) The contrasting effects of genome size, chromosome number and ploidy level on plant invasiveness: A global analysis. New Phytologist, 203, 697-703.
DOI PMID |
| [106] | Peng Y, Yang JX, Leitch IJ, Guignard MS, Seabloom EW, Cao D, Zhao FY, Li HL, Han XG, Jiang Y, Leitch AR, Wei CZ (2022) Plant genome size modulates grassland community responses to multi-nutrient additions. New Phytologist, 236, 2091-2102. |
| [107] |
Petrov DA (2001) Evolution of genome size: New approaches to an old problem. Trends in Genetics, 17, 23-28.
DOI PMID |
| [108] | Poggio L, Rosato M, Chiavarino AM, Naranjo CA (1998) Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Annals of Botany, 82, 107-115. |
| [109] | Price HJ (1988) Nuclear DNA content variation within angiosperm species. Evolutionary Trends in Plants, 2, 53-60. |
| [110] | Price HJ, Chambers KL, Bachmann K (1981) Geographic and ecological distribution of genomic DNA content variation in Microseris douglasii(Asteraceae). Botanical Gazette, 142, 415-426. |
| [111] |
Pyšek P, Skálová H, Čuda J, Guo WY, Suda J, Doležel J, Kauzál O, Lambertini C, Lučanová M, Mandáková T, Moravcová L, Pyšková K, Brix H, Meyerson LA (2018) Small genome separates native and invasive populations in an ecologically important cosmopolitan grass. Ecology, 99, 79-90.
DOI PMID |
| [112] | Rayburn AL (1990) Genome size variation in Southwestern United States Indian maize adapted to various altitudes. Evolutionary Trends in Plants, 4, 53-57. |
| [113] |
Rayburn AL, Auger JA (1990) Genome size variation in Zea mays ssp. mays adapted to different altitudes. Theoretical and Applied Genetics, 79, 470-474.
DOI PMID |
| [114] | Reeves G, Francis D, Davies MS, Rogers HJ, Hodkinson TR (1998) Genome size is negatively correlated with altitude in natural populations of Dactylis glomerata. Annals of Botany, 82, 99-105. |
| [115] | Rejmánek M (1996) A theory of seed plant invasiveness: The first sketch. Biological Conservation, 78, 171-181. |
| [116] | Renzaglia KS, Rasch EM, Pike LM (1995) Estimates of nuclear DNA content in bryophyte sperm cells: Phylogenetic considerations. American Journal of Botany, 82, 18-25. |
| [117] |
Roddy AB, Théroux-Rancourt G, Abbo T, Benedetti JW, Brodersen CR, Castro M, Castro S, Gilbride AB, Jensen B, Jiang GF, Perkins JA, Perkins SD, Loureiro J, Syed Z, Thompson RA, Kuebbing SE, Simonin KA (2020) The scaling of genome size and cell size limits maximum rates of photosynthesis with implications for ecological strategies. International Journal of Plant Sciences, 181, 75-87.
DOI |
| [118] | Schmitt MR, Edwards GE (1981) Photosynthetic capacity and nitrogen use efficiency of maize, wheat, and rice: A comparison between C3 and C4 photosynthesis. Journal of Experimental Botany, 32, 459-466. |
| [119] |
Shao C, Li YQ, Luo A, Wang ZH, Xi ZX, Liu JQ, Xu XT (2021) Relationship between functional traits and genome size variation of angiosperms with different life forms. Biodiversity Science, 29, 575-585. (in Chinese with English abstract)
DOI |
|
[ 邵晨, 李耀琪, 罗奥, 王志恒, 席祯翔, 刘建全, 徐晓婷 (2021) 不同生活型被子植物功能性状与基因组大小的关系. 生物多样性, 29, 575-585.]
DOI |
|
| [120] | Shi MJ, Cheng YY, Zhang WT, Xia XQ (2016) The evolutionary mechanism of genome size. Chinese Science Bulletin, 61, 3188-3195. (in Chinese) |
| [ 石米娟, 程莹寅, 张婉婷, 夏晓勤 (2016) 浅析基因组大小的进化机制. 科学通报, 61, 3188-3195.] | |
| [121] | Simonin KA, Roddy AB (2018) Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLoS Biology, 16, e2003706. |
| [122] | Šímová I, Herben T (2012) Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences, 279, 867-875. |
| [123] | Sims LE, Price HJ (1985) Nuclear DNA content variation in Helianthus(Asteraceae). American Journal of Botany, 72, 1213-1219. |
| [124] |
Singh KP, Raina SN, Singh AK (1996) Variation in chromosomal DNA associated with the evolution of Arachis species. Genome, 39, 890-897.
PMID |
| [125] | Šmarda P, Bureš P, Horová L, Foggi B, Rossi G (2008) Genome size and GC content evolution of Festuca: Ancestral expansion and subsequent reduction. Annals of Botany, 101, 421-433. |
| [126] | Šmarda P, Bureš P, Horová L, Leitch IJ, Mucina L, Pacini E, Tichý L, Grulich V, Rotreklová O (2014) Ecological and evolutionary significance of genomic GC content diversity in monocots. Proceedings of the National Academy of Sciences, USA, 111, E4096-E4102. |
| [127] |
Šmarda P, Hejcman M, Březinová A, Horová L, Steigerová H, Zedek F, Bureš P, Hejcmanová P, Schellberg J (2013) Effect of phosphorus availability on the selection of species with different ploidy levels and genome sizes in a long-term grassland fertilization experiment. New Phytologist, 200, 911-921.
DOI PMID |
| [128] | Souza G, Costa L, Guignard MS, Van-Lume B, Pellicer J, Gagnon E, Leitch IJ, Lewis GP (2019) Do tropical plants have smaller genomes? Correlation between genome size and climatic variables in the Caesalpinia Group (Caesalpinioideae, Leguminosae). Perspectives in Plant Ecology, Evolution and Systematics, 38, 13-23. |
| [129] | Sterner RW, Elser JJ (2002) Ecological Stoichiometry:The Biology of Elements from Molecules to the Biosphere. Princeton University Press, Princeton, New Jersey. |
| [130] | Suda J, Kyncl T, Freiova R (2003) Nuclear DNA amounts in Macronesian angiosperms. Annals of Botany, 92, 152-164. |
| [131] |
Suda J, Meyerson LA, Leitch IJ, Pyšek P (2015) The hidden side of plant invasions: The role of genome size. New Phytologist, 205, 994-1007.
DOI PMID |
| [132] |
Szarski H (1983) Cell size and the concept of wasteful and frugal evolutionary strategies. Journal of Theoretical Biology, 105, 201-209.
PMID |
| [133] |
Temsch EM, Greilhuber J (2001) Genome size in Arachis duranensis: A critical study. Genome, 44, 826-830.
PMID |
| [134] | Temsch EM, Temsch W, Ehrendorfer-Schratt L, Greilhuber J (2010) Heavy metal pollution, selection, and genome size: The species of the Žerjav study revisited with flow cytometry. Journal of Botany, 2010, 596542. |
| [135] | Teoh SB, Rees H (1976) Nuclear DNA amounts in populations of Picea and Pinus species. Heredity, 36, 123-137. |
| [136] | Théroux-Rancourt G, Roddy AB, Earles JM, Gilbert ME, Zwieniecki MA, Boyce CK, Tholen D, McElrone AJ, Simonin KA, Brodersen CR (2021) Maximum CO2 diffusion inside leaves is limited by the scaling of cell size and genome size. Proceedings of the Royal Society B: Biological Sciences, 288, 20203145. |
| [137] | Thibault J (1998) Nuclear DNA amount in pure species and hybrid willows (Salix): A flow cytometric investigation. Canadian Journal of Botany, 76, 157-165. |
| [138] | Torrel M, Vallès J (2001) Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): Systematic, evolutionary, and ecological implications. Genome, 44, 231-238. |
| [139] |
Turpeinen T, Kulmala J, Nevo E (1999) Genome size variation in Hordeum spontaneum populations. Genome, 42, 1094-1099.
PMID |
| [140] |
Van’t Hof J (1965) Relationships between mitotic cycle duration, S period duration and the average rate of DNA synthesis in the root meristem cells of several plants. Experimental Cell Research, 39, 48-58.
PMID |
| [141] | Van’t Hof J, Sparrow AH (1963) A relationship between DNA content, nuclear volume, and minimum mitotic cycle time. Proceedings of the National Academy of Sciences, USA, 49, 897-902. |
| [142] |
Veselý P, Bureš P, Šmarda P, Pavlíček T (2012) Genome size and DNA base composition of geophytes: The mirror of phenology and ecology? Annals of Botany, 109, 65-75.
DOI PMID |
| [143] |
Vidic T, Greilhuber J, Vilhar B, Dermastia M (2009) Selective significance of genome size in a plant community with heavy metal pollution. Ecological Applications, 19, 1515-1521.
PMID |
| [144] |
Vinogradov AE (1995) Nucleotypic effect in homeotherms: Body-mass-corrected basal metabolic rate of mammals is related to genome size. Evolution, 49, 1249-1259.
DOI PMID |
| [145] |
Vinogradov AE (1997) Nucleotypic effect in homeotherms: Body-mass independent resting metabolic rate of passerine birds is related to genome size. Evolution, 51, 220-225.
DOI PMID |
| [146] |
Vinogradov AE (2001) Mirrored genome size distributions in monocot and dicot plants. Acta Biotheoretica, 49, 43-51.
PMID |
| [147] |
Vinogradov AE (2003) Selfish DNA is maladaptive: Evidence from the plant Red List. Trends in Genetics, 19, 609-614.
DOI PMID |
| [148] | Wakamiya I, Newton RJ, Johnston SJ, Price JH (1993) Genome size and environmental factors in the genus Pinus. American Journal of Botany, 80, 1235-1241. |
| [149] |
Wang DD, Zheng ZY, Li Y, Hu HY, Wang ZY, Du X, Zhang SZ, Zhu MJ, Dong LW, Ren GP, Yang YZ (2021) Which factors contribute most to genome size variation within angiosperms? Ecology and Evolution, 11, 2660-2668.
DOI PMID |
| [150] |
Wang J, Chen P, Wang GJ, Keller L (2010) Chromosome size differences may affect meiosis and genome size. Science, 329, 293.
DOI PMID |
| [151] | Zhao FY, Wei CZ, Lü XT, Han XG (2019) Responses of plant species with different genome size to water and nitrogen addition in Inner Mongolia Grassland, China. Chinese Journal of Applied Ecology, 30, 2675-2681. (in Chinese with English abstract) |
|
[ 赵芳媛, 魏存争, 吕晓涛, 韩兴国 (2019) 内蒙古草原不同基因组大小植物对氮水添加的响应. 应用生态学报, 30, 2675-2681.]
DOI |
|
| [152] | Zonneveld BJM, Duncan GD (2006) Genome size for the species of Nerine Herb. (Amaryllidaceae) and its evident correlation with growth cycle, leaf width and other morphological characters. Plant Systematics and Evolution, 257, 251-260. |
| [1] | 王瑞武, 于云云, 朱其凯, 王超, 李敏岚, 韩嘉旭. 路径依赖的选择——统一自然选择与中性选择[J]. 生物多样性, 2024, 32(7): 24120-. |
| [2] | 金恒镳. 从天择到人择: 在华莱士的肩膀上看地球的未来[J]. 生物多样性, 2023, 31(12): 23267-. |
| [3] | 高德, 王彦平. 小岛屿效应检测方法研究进展[J]. 生物多样性, 2023, 31(12): 23299-. |
| [4] | 王瑞武, 李敏岚, 韩嘉旭, 王超. 适合度的相对性与路径依赖的自然选择[J]. 生物多样性, 2022, 30(1): 21323-. |
| [5] | 邵晨, 李耀琪, 罗奥, 王志恒, 席祯翔, 刘建全, 徐晓婷. 不同生活型被子植物功能性状与基因组大小的关系[J]. 生物多样性, 2021, 29(5): 575-585. |
| [6] | 董雷, 王静, 刘永刚, 赵志平, 米湘成, 郭柯. 太行山北段地区荆条灌丛和三裂绣线菊灌丛群落谱系结构[J]. 生物多样性, 2021, 29(1): 21-31. |
| [7] | 李忠虎, 刘占林, 王玛丽, 钱增强, 赵鹏, 祝娟, 杨一欣, 阎晓昊, 李银军, 赵桂仿. 基因流存在条件下的物种形成研究述评:生殖隔离机制进化[J]. 生物多样性, 2014, 22(1): 88-96. |
| [8] | 赵紫华, 王颖, 贺达汉, 张蓉, 朱猛蒙, 董风林. 苜蓿草地生境丧失与破碎化对昆虫物种丧失与群落重建的影响[J]. 生物多样性, 2011, 19(4): 453-462. |
| [9] | 孙海芹, 李昂, 班玮, 郑晓明, 葛颂. 濒危植物独花兰的形态变异及其适应意义[J]. 生物多样性, 2005, 13(5): 376-386. |
| [10] | 尹玉峰, 王昊, 陈艾, 刘国琪. 对大熊猫数量调查方法中咬节区分机制的准确性评价[J]. 生物多样性, 2005, 13(5): 439-444. |
| [11] | 刘占林, 赵桂仿. 居群遗传学原理及其在珍稀濒危植物保护中的应用[J]. 生物多样性, 1999, 07(4): 340-346. |
| [12] | 张大勇, 姜新华. 遗传多样性与濒危植物保护生物学研究进展[J]. 生物多样性, 1999, 07(1): 31-37. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()