



生物多样性 ›› 2022, Vol. 30 ›› Issue (2): 21258. DOI: 10.17520/biods.2021258 cstr: 32101.14.biods.2021258
宋佳1, 职铭阳1, 陈强2, 李玥莹1, 吴隆坤3, 农保选4, 李丹婷4, 逄洪波1,*( ), 郑晓明5,*(
), 郑晓明5,*( )
)
收稿日期:2021-06-29
接受日期:2021-10-14
出版日期:2022-02-20
发布日期:2022-02-28
通讯作者:
逄洪波,郑晓明
作者简介:zhengxiaoming@caas.cn基金资助:
Jia Song1, Mingyang Zhi1, Qiang Chen2, Yueying Li1, Longkun Wu3, Baoxuan Nong4, Danting Li4, Hongbo Pang1,*( ), Xiaoming Zheng5,*(
), Xiaoming Zheng5,*( )
)
Received:2021-06-29
Accepted:2021-10-14
Online:2022-02-20
Published:2022-02-28
Contact:
Hongbo Pang,Xiaoming Zheng
摘要:
为探究CTB4a基因的自然变异是否与栽培稻苗期耐低温相关, 本研究以133份栽培稻(Oryza sativa L.)及35份普通野生稻(O. rufipogon Griff.)为实验材料, 对CTB4a编码区的核苷酸多样性及其单倍型与地理分布的关系进行分析。结果表明CTB4a基因编码区有14个核苷酸变异, 组成33个单倍型。Network分析发现, 这些单倍型的286 bp处的SNP变异(+2,035,097 bp, G > C; +96 aa, Ala→Pro)将其分成Group A和Group B两组。Group A (CTB4ajap)共有56份样品, 其中有43份(76.79%)是种植于中高纬度地区的粳稻; Group B (CTB4aind)有77份样品, 有63份(83.12%)是种植于热带和亚热带国家的籼稻。Group A中样品苗期黄叶率的平均值比Group B样品低30%, 且两者耐寒性具有显著差异(P = 1.25e-09)。位于286 bp处的变异只在Group A中出现, 在Group B中均不存在, 说明随着水稻种植区域向北迁移的育种过程中, 栽培稻中出现了CTB4ajap并被固定下来。该研究为理解水稻对低温适应的分子遗传机制和培育耐寒品种提供了理论基础。
宋佳, 职铭阳, 陈强, 李玥莹, 吴隆坤, 农保选, 李丹婷, 逄洪波, 郑晓明 (2022) 水稻耐寒基因CTB4a的核苷酸多样性及区域适应性. 生物多样性, 30, 21258. DOI: 10.17520/biods.2021258.
Jia Song, Mingyang Zhi, Qiang Chen, Yueying Li, Longkun Wu, Baoxuan Nong, Danting Li, Hongbo Pang, Xiaoming Zheng (2022) Nucleotide diversity and adaptation of CTB4a gene related to cold tolerance in rice. Biodiversity Science, 30, 21258. DOI: 10.17520/biods.2021258.

图1 CTB4a基因编码区核苷酸多样性。A: 133份栽培稻CTB4a基因编码区核苷酸多样性; B: CTB4a基因编码的氨基酸序列及其结构域, 灰色部分显示的是LRR-RLK结构域; C: 35份野生稻编码区的核苷酸多样性。图A和C中灰色部分展示的是氨基酸的变异位点(SNP和InDel)及Group A和Group B两组中籼稻群体平均黄叶率数值; 图A和C首列分别表示CTB4a基因33个单倍型和33个野生稻的名称。cvs: 品种; LYR: 黄叶率; I: 籼稻; J: 粳稻。
Fig. 1 Nucleotide diversity in the CTB4a coding region. A, Nucleotide diversity in the coding region of CTB4a gene in 133 cultivated rice samples; B, Amino acid sequence and domains encoded by CTB4a gene, the gray part shows the LRR-RLK domain; C, Nucleotide diversity in the Coding Region of 35 wild rice. Amino acid variation sites in gray. In A and C, Amino acid variation sites (SNP and InDel) and average yellow leaf rate (LYR) values of indica rice population in two groups were shown in gray. And the first column of A and C shows the names of 33 haplotypes of CTB4a gene and 33 wild rice, respectively. cvs, Cultivars; LYR, Yellow leaf rate; I, O. sativa ssp. indica; J, O. sativa ssp. japonica.
| 元件名称 Name | 元件功能 Function | 元件存在类型 Existence type | 元件数量 Number |
|---|---|---|---|
| LTR | 低温诱导 Low-temperature induction | 耐寒/不耐寒 Cold tolerance/intolerance | 1 |
| ABRE | 脱落酸诱导 Abscisic acid induction | 3 | |
| AE-box | 光诱导 Light induction | 10 | |
| G-box | |||
| Box 4 | |||
| Box II | |||
| GATA-motif | |||
| TCCC-motif | |||
| TCT-motif | |||
| ACA-motif | 耐寒 Cold tolerance | 1 | |
| MBS | 干旱诱导 Drought induction | 不耐寒 Cold intolerance | 1 |
表1 CTB4a基因启动子顺式作用元件分析
Table 1 Cis-acting elements analysis of CTB4a gene promoter
| 元件名称 Name | 元件功能 Function | 元件存在类型 Existence type | 元件数量 Number |
|---|---|---|---|
| LTR | 低温诱导 Low-temperature induction | 耐寒/不耐寒 Cold tolerance/intolerance | 1 |
| ABRE | 脱落酸诱导 Abscisic acid induction | 3 | |
| AE-box | 光诱导 Light induction | 10 | |
| G-box | |||
| Box 4 | |||
| Box II | |||
| GATA-motif | |||
| TCCC-motif | |||
| TCT-motif | |||
| ACA-motif | 耐寒 Cold tolerance | 1 | |
| MBS | 干旱诱导 Drought induction | 不耐寒 Cold intolerance | 1 |
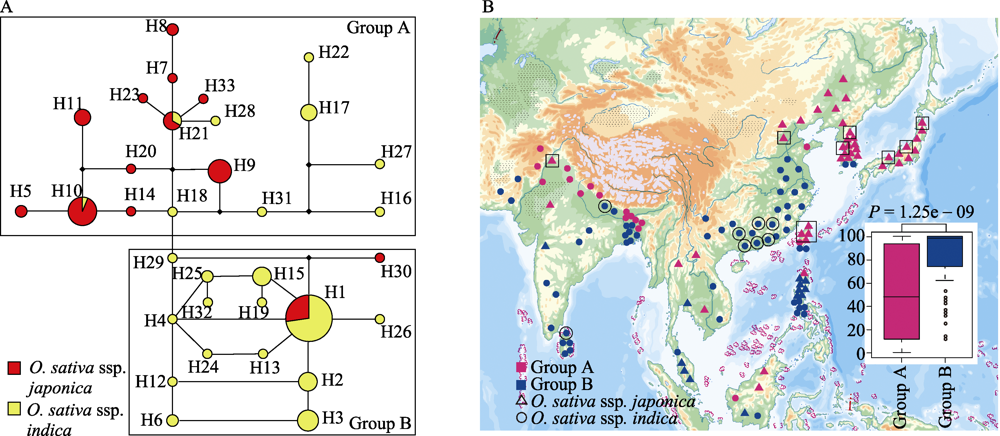
图2 CTB4a基因编码区单倍型的Network图(A)和栽培稻地理分布图(B)。图A中红色表示粳稻, 黄色表示籼稻。图B中粉色表示Group A, 蓝色表示Group B; 三角形代表粳稻, 圆形代表籼稻; 方框圈出为H9单倍型, 圆形圈出为H3单倍型。
Fig. 2 Network of haplotypes of CTB4a coding region (A) and geographic distribution map in cultivated rice (B). A, The network of all haplotypes for CTB4a, O. sativa ssp. japonica in red and O. sativa ssp. indica in yellow; B, Geographic distribution map of cultivated rice in group A and group B. Group A in pink and Group B in blue; O. sativa ssp. japonica with triangle and O. sativa ssp. indica with circle; H9 haplotype in square; H3 haplotype in circle.
| [1] |
Andaya VC, Mackill DJ (2003a) Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. Journal of Experimental Botany, 54, 2579-2585.
DOI URL |
| [2] |
Andaya VC, Mackill DJ (2003b) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theoretical and Applied Genetics, 106, 1084-1090.
DOI URL |
| [3] |
Chang TT (1976) The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica, 25, 425-441.
DOI URL |
| [4] |
Chory J, Wu DY (2001) Weaving the complex web of signal transduction. Plant Physiology, 125, 77-80.
PMID |
| [5] |
Conley TR, Park SC, Kwon HB, Peng HP, Shih MC (1994) Characterization of cis-acting elements in light regulation of the nuclear gene encoding the A subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Molecular and Cellular Biology, 14, 2525-2533.
DOI PMID |
| [6] |
Cruz RPD, Sperotto RA, Cargnelutti D, Adamski JM, FreitasTerra T, Fett JP (2013) Avoiding damage and achieving cold tolerance in rice plants. Food and Energy Security, 2, 96-119.
DOI URL |
| [7] |
Ding YL, Shi YT, Yang SH (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytologist, 222, 1690-1704.
DOI URL |
| [8] |
Guo HF, Zeng YW, Li JL, Ma XQ, Zhang ZY, Lou QJ, Li J, Gu YS, Zhang HL, Li JJ, Li ZC (2020) Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnology Journal, 18, 2491-2503.
DOI URL |
| [9] |
Guo XY, Liu DF, Chong K (2018) Cold signaling in plants: Insights into mechanisms and regulation. Journal of Integrative Plant Biology, 60, 745-756.
DOI URL |
| [10] |
Hedrick PW (2013) Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology, 22, 4606-4618.
DOI PMID |
| [11] |
Huang CL, Hung CY, Chiang YC, Hwang CC, Hsu TW, Huang CC, Hung KH, Tsai KC, Wang KH, Osada N, Schaal BA, Chiang TY (2012) Footprints of natural and artificial selection for photoperiod pathway genes in Oryza. The Plant Journal, 70, 769-782.
DOI URL |
| [12] |
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology, 35, 25-34.
PMID |
| [13] |
Kim SI, Andaya VC, Tai TH (2011) Cold sensitivity in rice (Oryza sativa L.) is strongly correlated with a naturally occurring I99V mutation in the multifunctional glutathione transferase isoenzyme GSTZ2. The Biochemical Journal, 435, 373-380.
DOI URL |
| [14] |
Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends in Genetics, 23, 578-587.
DOI URL |
| [15] |
Li JL, Zeng YW, Pan YH, Zhou L, Zhang ZY, Guo HF, Lou QJ, Shui GH, Huang HG, Tian H, Guo YM, Yuan PR, Yang H, Pan GJ, Wang RY, Zhang HL, Yang SH, Guo Y, Ge S, Li JJ, Li ZC (2021) Stepwise selection of natural variations at CTB2 and CTB4a improves cold adaptation during domestication of japonica rice. New Phytologist, 231, 1056-1072.
DOI URL |
| [16] |
Liu CT, Ou SJ, Mao BG, Tang JY, Wang W, Wang HR, Cao SY, Schläppi MR, Zhao BR, Xiao GY, Wang XP, Chu CC (2018) Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nature Communications, 9, 3302.
DOI URL |
| [17] |
Liu FX, Sun CQ, Tan LB, Fu YC, Li DJ, Wang XK (2003) Identification and mapping of quantitative trait loci controlling cold-tolerance of Chinese common wild rice (O. rufipogon Griff.) at booting to flowering stages. Chinese Science Bulletin, 48, 2068-2071.
DOI URL |
| [18] |
Lv Y, Hussain MA, Luo D, Tang N (2019) Current understanding of genetic and molecular basis of cold tolerance in rice. Molecular Breeding, 39, 159.
DOI URL |
| [19] |
Ma Y, Dai XY, Xu YY, Luo W, Zheng XM, Zeng DL, Pan YJ, Lin XL, Liu HH, Zhang DJ, Xiao J, Guo XY, Xu SJ, Niu YD, Jin JB, Zhang H, Xu X, Li LG, Wang W, Qian Q, Ge S, Chong K (2015) COLD1 confers chilling tolerance in rice. Cell, 160, 1209-1221.
DOI URL |
| [20] | Mao DH, Xin YY, Tan YJ, Hu XJ, Bai JJ, Liu ZY, Yu YL, Li LY, Peng C, Fan T, Zhu YX, Guo YL, Wang SH, Lu DP, Xing YZ, Yuan LP, Chen CY (2019) Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proceedings of the National Academy of Sciences, USA, 116, 3494-3501. |
| [21] |
Mazzella MA, Bertero D, Casal JJ (2000) Temperature- dependent internode elongation in vegetative plants of Arabidopsis thaliana lacking phytochrome B and cryptochrome 1. Planta, 210, 497-501.
PMID |
| [22] | Oka HI (1958) Intervarietal variation and classification of cultivated rice. Indian Journal of Genetics Plant Breeding, 18, 79-89. |
| [23] |
Penfield S (2008) Temperature perception and signal transduction in plants. New Phytologist, 179, 615-628.
DOI PMID |
| [24] |
Roy S, Choudhury SR, Singh SK, Das KP (2012) Functional analysis of light-regulated promoter region of AtPolλ gene. Planta, 235, 411-432.
DOI URL |
| [25] |
Saito K, Hayano-Saito Y, Kuroki M, Sato Y (2010) Map-based cloning of the rice cold tolerance gene Ctb1. Plant Science, 179, 97-102.
DOI URL |
| [26] |
Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Current Opinion in Plant Biology, 3, 138-142.
PMID |
| [27] |
Shirasawa S, Endo T, Nakagomi K, Yamaguchi M, Nishio T (2012) Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar, ‘Lijiangxin tuanheigu’, in rice, Oryza sativa L. Theoretical and Applied Genetics, 124, 937-946.
DOI PMID |
| [28] |
Sthapit BR, Witcombe JR (1998) Inheritance of tolerance to chilling stress in rice during germination and plumule greening. Crop Science, 38, 660-665.
DOI URL |
| [29] |
Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R (1998) Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA, 4, 1549-1568.
PMID |
| [30] |
Xiao N, Gao Y, Qian HJ, Gao Q, Wu YY, Zhang DP, Zhang XX, Yu L, Li YH, Pan CH, Liu GQ, Zhou CH, Jiang M, Huang NS, Dai ZY, Liang CZ, Chen Z, Chen JM, Li AH (2018) Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiology, 177, 1108-1123.
DOI URL |
| [31] |
Xie GS, Kato H, Imai R (2012) Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. The Biochemical Journal, 443, 95-102.
DOI URL |
| [32] |
Zhang ZY, Li JJ, Pan YH, Li JL, Zhou L, Shi HL, Zeng YW, Guo HF, Yang SM, Zheng WW, Yu JP, Sun XM, Li GL, Ding YL, Ma L, Shen SQ, Dai LY, Zhang HL, Yang SH, Guo Y, Li ZC (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nature Communications, 8, 14788.
DOI URL |
| [33] |
Zhao J, Wang SS, Qin JJ, Sun CQ, Liu FX (2020) The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnology Journal, 18, 756-769.
DOI PMID |
| [34] |
Zhao JL, Zhang SH, Dong JF, Yang TF, Mao XX, Liu Q, Wang XF, Liu B (2017) A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnology Journal, 15, 1141-1148.
DOI URL |
| [1] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [2] | 谢华, 杨培, 李宗波. 鸡嗉子榕传粉榕小蜂表皮碳氢化合物的性二型及季节变化[J]. 生物多样性, 2024, 32(6): 24001-. |
| [3] | 杨锐, 侯姝彧, 张引, 赵智聪. 论建立中国自然保护兼用地的必要性和可行性[J]. 生物多样性, 2024, 32(4): 23454-. |
| [4] | 景昭阳, 程可光, 舒恒, 马永鹏, 刘平丽. 全基因组重测序方法在濒危植物保护中的应用[J]. 生物多样性, 2023, 31(5): 22679-. |
| [5] | 邵雯雯, 范国祯, 何知舟, 宋志平. 多地同质园实验揭示普通野生稻的表型可塑性与本地适应性[J]. 生物多样性, 2023, 31(3): 22311-. |
| [6] | 罗瑞, 陈娅, 张汉马. 芸薹属植物全基因组重测序研究进展[J]. 生物多样性, 2023, 31(10): 23237-. |
| [7] | 胡正艳, 郑全晶, 母其勇, 杜志强, 刘琳, 星耀武, 韩廷申. 不同纬度高蔊菜的交配系统和繁殖保障[J]. 生物多样性, 2021, 29(6): 712-721. |
| [8] | 王婷, 夏增强, 舒江平, 张娇, 王美娜, 陈建兵, 王慷林, 向建英, 严岳鸿. 全基因组复制事件的绝对定年揭示莲座蕨属植物的迟滞演化[J]. 生物多样性, 2021, 29(6): 722-734. |
| [9] | 王宇飞,苏红巧,赵鑫蕊,苏杨,罗敏. 基于保护地役权的自然保护地适应性管理方法探讨: 以钱江源国家公园体制试点区为例[J]. 生物多样性, 2019, 27(1): 88-96. |
| [10] | 宋志平, 陈家宽, 赵耀. 水稻驯化与长江文明[J]. 生物多样性, 2018, 26(4): 346-356. |
| [11] | 刘莉, 舒江平, 韦宏金, 张锐, 沈慧, 严岳鸿. 东亚特有珍稀蕨类植物岩穴蕨(碗蕨科)高通量转录组测序及分析[J]. 生物多样性, 2016, 24(12): 1325-1334. |
| [12] | 许强华, 吴智超, 陈良标. 南极鱼类多样性和适应性进化研究进展[J]. 生物多样性, 2014, 22(1): 80-87. |
| [13] | 王雨, 叶又茵, 林茂, 陈兴群. 南海北部夜光藻种群的时空分布及其环境适应性[J]. 生物多样性, 2012, 20(6): 685-692. |
| [14] | 田中平, 庄丽, 李建贵, 程模香. 伊犁河谷北坡野果林木本植物种间关系 及环境解释[J]. 生物多样性, 2011, 19(3): 335-342. |
| [15] | 严岳鸿, 何祖霞, 苑虎, 邢福武. 坡向差异对广东古兜山自然保护区蕨类植物多样性的生态影响[J]. 生物多样性, 2011, 19(1): 41-47. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn