



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (4): 414-419. DOI: 10.3724/SP.J.1003.2010.414 cstr: 32101.14.SP.J.1003.2010.414
Special Issue: 传粉生物学; 昆虫多样性与生态功能
• Special Issue • Previous Articles Next Articles
Yiyi Long, Liyuan Yang, Wanjin Liao( )
)
Received:2010-04-22
Accepted:2010-06-26
Online:2010-07-20
Published:2010-07-20
Contact:
Wanjin Liao
Yiyi Long, Liyuan Yang, Wanjin Liao. Identifying cryptic species in pollinating-fig wasps by PCR-RFLP on mtDNA COI gene[J]. Biodiv Sci, 2010, 18(4): 414-419.
| 样品编号 Sample No. | 大果榕小蜂 C. emarginatus | 鸡嗉果榕小蜂 C. gravelyi | |||
|---|---|---|---|---|---|
| 隐种A头数 Number of A | 隐种B头数Number of B | 隐种A头数 Number of A | 隐种B头数 Number of B | ||
| 1 | 1 | 10 | 6 | 2 | |
| 2 | 6 | 2 | 10 | 2 | |
| 3 | 10 | 2 | 20 | 2 | |
| 4 | 20 | 2 | 1 | 10 | |
| 5 | 2 | 20 | 2 | 20 | |
| 6 | 2 | 10 | 2 | 10 | |
| 7 | 2 | 6 | 2 | 6 | |
| 8 | 4 | 4 | 4 | 4 | |
| 9 | 0 | 2 | 0 | 2 | |
| 10 | 2 | 0 | 2 | 0 | |
Table 1 Composition of cryptic species (A and B) in Ceratosolen emarginatus and C. gravelyi used in this study
| 样品编号 Sample No. | 大果榕小蜂 C. emarginatus | 鸡嗉果榕小蜂 C. gravelyi | |||
|---|---|---|---|---|---|
| 隐种A头数 Number of A | 隐种B头数Number of B | 隐种A头数 Number of A | 隐种B头数 Number of B | ||
| 1 | 1 | 10 | 6 | 2 | |
| 2 | 6 | 2 | 10 | 2 | |
| 3 | 10 | 2 | 20 | 2 | |
| 4 | 20 | 2 | 1 | 10 | |
| 5 | 2 | 20 | 2 | 20 | |
| 6 | 2 | 10 | 2 | 10 | |
| 7 | 2 | 6 | 2 | 6 | |
| 8 | 4 | 4 | 4 | 4 | |
| 9 | 0 | 2 | 0 | 2 | |
| 10 | 2 | 0 | 2 | 0 | |
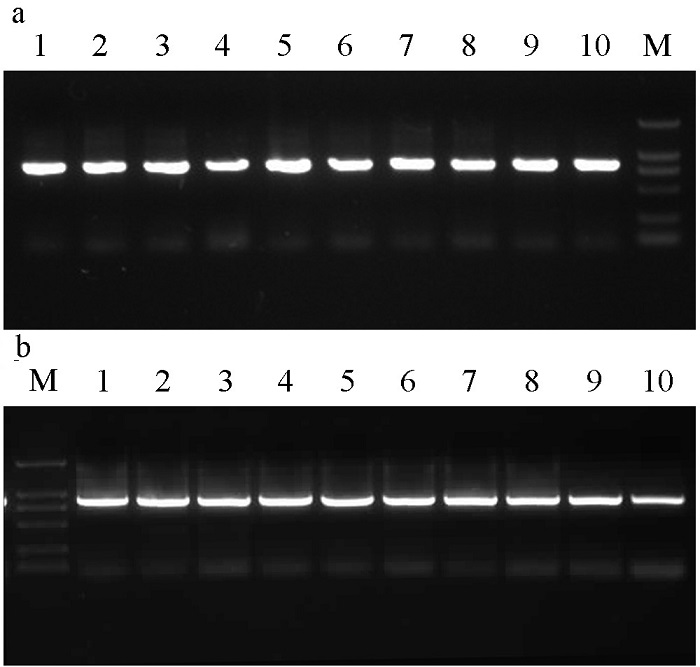
Fig. 1 The PCR products of COI gene in Ceratosolen emarginatus (A) and C. gravelyi (B). M indicates the DNA marker, and the numbers indicate the sample number. See Table 1 for the composition of cryptic species for each sample.
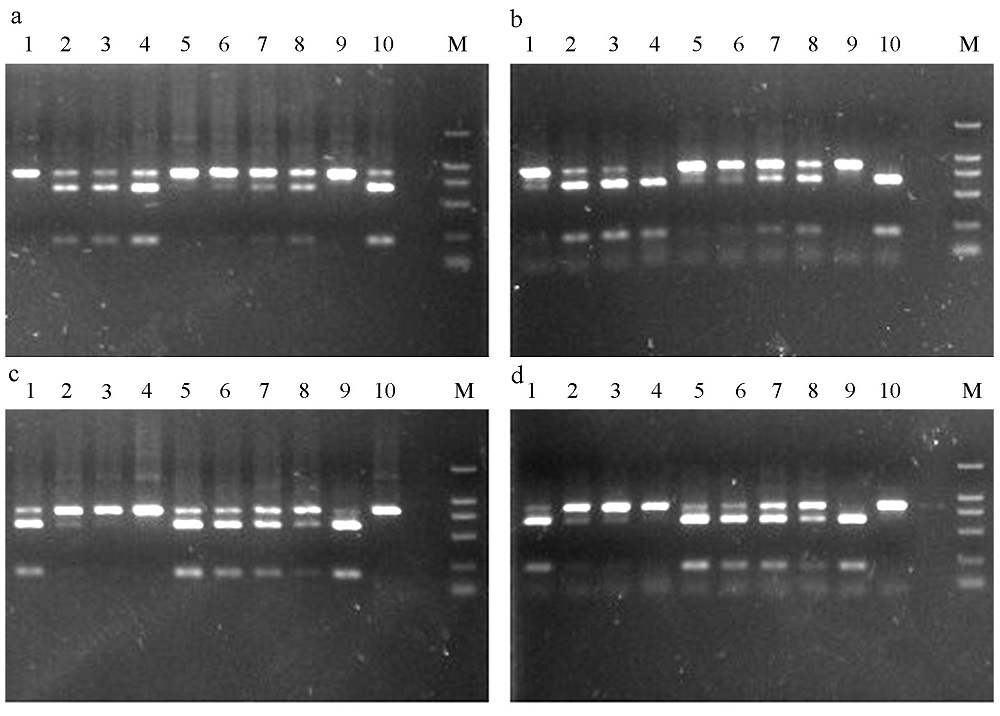
Fig. 2 The digested products by XhoI (a and b) and BssSI (c and d) in Ceratosolen emarginatus. a and c show the digested results of purified PCR products; b and d, unpurified. M is the DNA marker (DL 2000). 1-10 correspond to the sample numbers in Table 1.
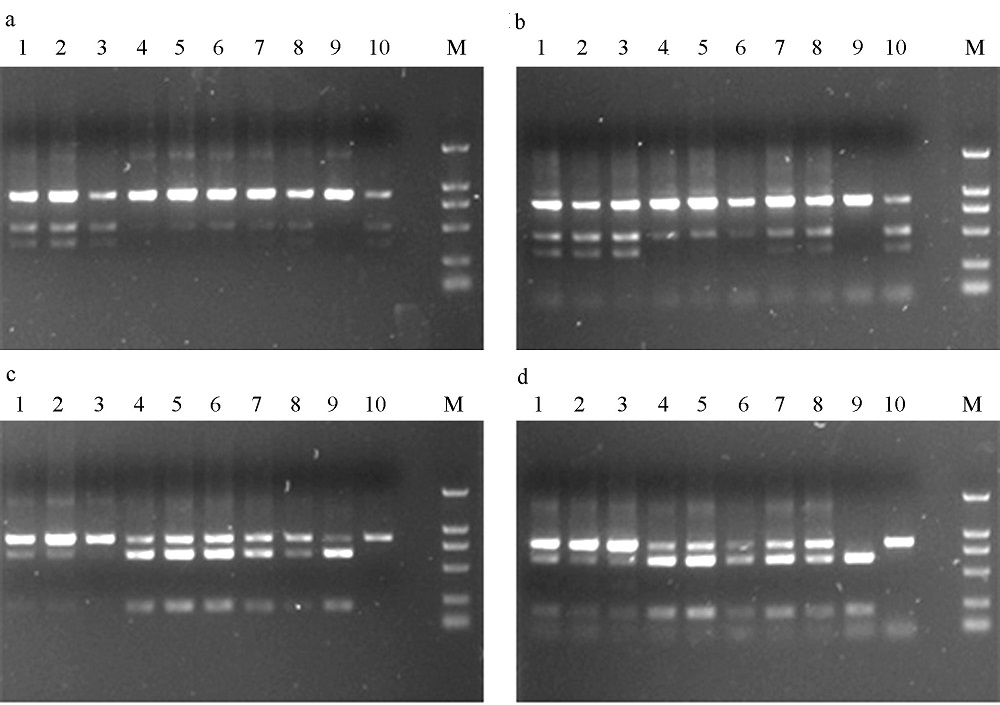
Fig. 3 The digested products by BmrI (a and b) and AvaI (c and d) in Ceratosolen gravelyi. a and c show the digested results of purified PCR products; b and d, unpurified. 1-10 correspond to the sample numbers in Table 1.
| [1] |
Brown GG, Gadaleta G, Pepe G, Saccone C (1986) Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. Journal of Molecular Biology, 192,503-511.
DOI URL PMID |
| [2] |
Chen SY, Su YH, Wu SF, Sha T, Zhang YP (2005) Mitochondrial diversity and phylogeographic structure of Chinese domestic goats. Molecular Phylogenetics and Evolution, 37,804-814.
DOI URL PMID |
| [3] | Cook JM, Rasplus JY (2003) Mutualists with attitude: coevolving fig wasps and figs. Trends in Ecology and Evolution, 18,241-248. |
| [4] |
Cook JM, Segar ST (2010) Speciation in fig wasps. Ecological Entomology, 35,54-66.
DOI URL |
| [5] | Fajardo V, Gonzalez I, Dooley J, Garret S, Brown HM, Garcia T, Martin R (2009) Application of polymerase chain reaction-restriction fragment length polymorphism analysis and lab-on-a-chip capillary electrophoresis for the specific identification of game and domestic meats. Journal of the Science of Food and Agriculture, 89,843-847. |
| [6] |
Fajardo V, Gonzalez I, Lopez-Calleja I, Martin I, Rojas M, Hernandez PE, Garcia T, Martin R (2007) Identification of meats from red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) using polymerase chain reaction targeting specific sequences from the mitochondrial 12S rRNA gene. Meat Science, 76,234-240.
URL PMID |
| [7] |
Haine ER, Martin J, Cook JM (2006) Deep mtDNA divergences indicate cryptic species in a fig-pollinating wasp. BMC Evolutionary Biology, 6,83.
URL PMID |
| [8] | Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41,95-98. |
| [9] |
Karl SA, Bowen BW, Avise JC (1992) Global population genetic-structure and male-mediated gene flow in the green turtle (Chelonia mydas): RFLP analyses of anonymous nuclear loci. Genetics, 131,163-173.
URL PMID |
| [10] | Machado CA, Robbins N, Gilbert MTP, Herre EA (2005) Critical review of host specificity and its coevolutionary implications in the fig/fig-wasp mutualism. Proceedings of the National Academy of Sciences, USA, 102,6558-6565. |
| [11] | Mayr E (1942) Systematics and the Origin of Species. Columbia University Press, New York. |
| [12] |
Molbo D, Machado CA, Sevenster JG, Keller L, Herre EA (2003) Cryptic species of fig-pollinating wasps: implications for the evolution of the fig-wasp mutualism, sex allocation, and precision of adaptation. Proceedings of the National Academy of Sciences, USA, 100,5867-5872.
DOI URL |
| [13] | Peng YQ (彭艳琼), Yang DR (杨大荣), Zhou F (周芳), Zhang GM (张光明), Song QS (宋启示) (2003) Pollination biology of Ficus auriculata Lour. in tropical rainforest of Xishu- angbanna. Acta Phytoecologica Sinica (植物生态学报), 27,111-117. (in Chinese with English abstract) |
| [14] | Sharma N, Thind SS, Girish PS, Sharma D (2008) PCR-RFLP of 12S rRNA gene for meat speciation. Journal of Food Science and Technology-Mysore, 45,353-355. |
| [15] | Simon C, Frati F, Beckenback A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved PCR primers. Annals of the Entomological Society of America, 87,651-701. |
| [16] |
Van der Kuyl AC, Kuiken CL, Dekker JT, Goudsmit J (1995) Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. Journal of Molecular Evolution, 40,173-180.
DOI URL PMID |
| [17] |
Wang Q, Zhang X, Zhang HY, Zhang J, Chen GQ, Zhao DH, Ma HP, Liao WJ (2010) Identification of 12 animal species meat by T-RFLP on the 12S rRNA gene. Meat Science, 85,265-269.
DOI URL PMID |
| [18] | Wang QY (王秋艳), Yang DR (杨大荣), Peng YQ (彭艳琼) (2003) Pollination behaviour and propagation of pollinator wasps on Ficus semicordata in Xishuangbanna, China. Acta Entomologica Sinica (昆虫学报), 46,27-34. (in Chinese with English abstract) |
| [19] |
Weiblen GD (2002) How to be a fig wasp. Annual Review of Entomology, 47,299-330.
DOI URL PMID |
| [20] | Wiebes JT (1979) Co-evolution of figs and their insect pollinators. Annual Review of Entomology, 10,1-12. |
| [21] | Zou YP (邹喻苹), Ge S (葛颂), Wang XD (王晓东) (2001) Molecular Markers in Systematic and Evolutionary Plant Biology (系统与进化植物学中的分子标记). Science Press, Beijing. (in Chinese) |
| [1] | Jiajia Pu, Pingjun Yang, Yang Dai, Kexin Tao, Lei Gao, Yuzhou Du, Jun Cao, Xiaoping Yu, Qianqian Yang. Species identification and population genetic structure of non-native apple snails (Ampullariidea: Pomacea) in the lower reaches of the Yangtze River [J]. Biodiv Sci, 2023, 31(3): 22346-. |
| [2] | Keyi Wu, Wenda Ruan, Difeng Zhou, Qingchen Chen, Chengyun Zhang, Xinyuan Pan, Shang Yu, Yang Liu, Rongbo Xiao. Syllable clustering analysis-based passive acoustic monitoring technology and its application in bird monitoring [J]. Biodiv Sci, 2023, 31(1): 22370-. |
| [3] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [4] | Yuanyuan Li, Chaonan Liu, Rong Wang, Shuixing Luo, Shouqian Nong, Jingwen Wang, Xiaoyong Chen. Applications of molecular markers in conserving endangered species [J]. Biodiv Sci, 2020, 28(3): 367-375. |
| [5] | Jiaxin Kong, Zhaochen Zhang, Jian Zhang. Classification and identification of plant species based on multi-source remote sensing data: Research progress and prospect [J]. Biodiv Sci, 2019, 27(7): 796-812. |
| [6] | Weng Zhuoxian, Huang Jiaqiong, Zhang Shihao, Yu Kaichun, Zhong Fusheng, Huang Xunhe, Zhang Bin. Genetic diversity and population structure of black-bone chickens in China revealed by mitochondrial COI gene sequences [J]. Biodiv Sci, 2019, 27(6): 667-676. |
| [7] | Jianfeng Huang,Rui Xu,Yanqiong Peng. Progress on the breakdown of one-to-one rule in symbiosis of figs and their pollinating wasps [J]. Biodiv Sci, 2018, 26(3): 295-303. |
| [8] | Yun Cao, Wenjing Shen, Lian Chen, Feilong Hu, Lei Zhou, Haigen Xu. Application of metabarcoding technology in studies of fungal diversity [J]. Biodiv Sci, 2016, 24(8): 932-939. |
| [9] | Tai Wang, Yanping Zhang, Lihong Guan, Yanyan Du, Zhongyu Lou, Wenlong Jiao. Current freshwater fish resources and the application of DNA barcoding in species identification in Gansu Province [J]. Biodiv Sci, 2015, 23(3): 306-313. |
| [10] | Dawei Guan, Li Li, Xin Jiang, Mingchao Ma, Fengming Cao, Baoku Zhou, Jun Li. Influence of long-term fertilization on the community structure and diversity of soybean rhizobia in black soil [J]. Biodiv Sci, 2015, 23(1): 68-78. |
| [11] | Dangni Zhang, Lianming Zheng, Jinru He, Wenjing Zhang, Yuanshao Lin, Yang Li. DNA barcoding of hydromedusae in northern Beibu Gulf for species identification [J]. Biodiv Sci, 2015, 23(1): 50-60. |
| [12] | Chaolun Li, Minxiao Wang, Fangping Cheng, Song Sun. DNA barcoding and its application to marine zooplankton ecology [J]. Biodiv Sci, 2011, 19(6): 805-814. |
| [13] | Haifeng Gu, Tingting Liu, Dongzhao Lan. Progress of dinoflagellate cyst research in the China seas [J]. Biodiv Sci, 2011, 19(6): 779-786. |
| [14] | Yan Liu, Jianxiu Wang, Xuejun Ge, Tong Cao. The rps4 locus as an alternative marker for barcoding bryophytes: eva- luation based on data mining from GenBank [J]. Biodiv Sci, 2011, 19(3): 311-318. |
| [15] | Lin Xu, Jiajie Xu, Qiaoli Liu, Ruimei Xie, Gehong Wei. Genetic diversity in rhizobia isolated from Sphaerophysa salsula in several regions of northwestern China [J]. Biodiv Sci, 2009, 17(1): 69-75. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()