



Biodiv Sci ›› 2018, Vol. 26 ›› Issue (12): 1318-1324. DOI: 10.17520/biods.2018184 cstr: 32101.14.biods.2018184
• Original Papers: Microbial Diversity • Previous Articles Next Articles
Xiaojuan Deng1, Jianli Liu1, Xingfu Yan1, Peigui Liu2,*( )
)
Received:2018-07-03
Accepted:2018-12-21
Online:2018-12-20
Published:2019-02-11
Contact:
Liu Peigui
About author:# 同等贡献作者 Contributed equally to this work
Xiaojuan Deng, Jianli Liu, Xingfu Yan, Peigui Liu. Community composition of bacteria associated with ascocarps of Tuber indicum using traditional culture method and Roche 454 high-throughput sequencing[J]. Biodiv Sci, 2018, 26(12): 1318-1324.
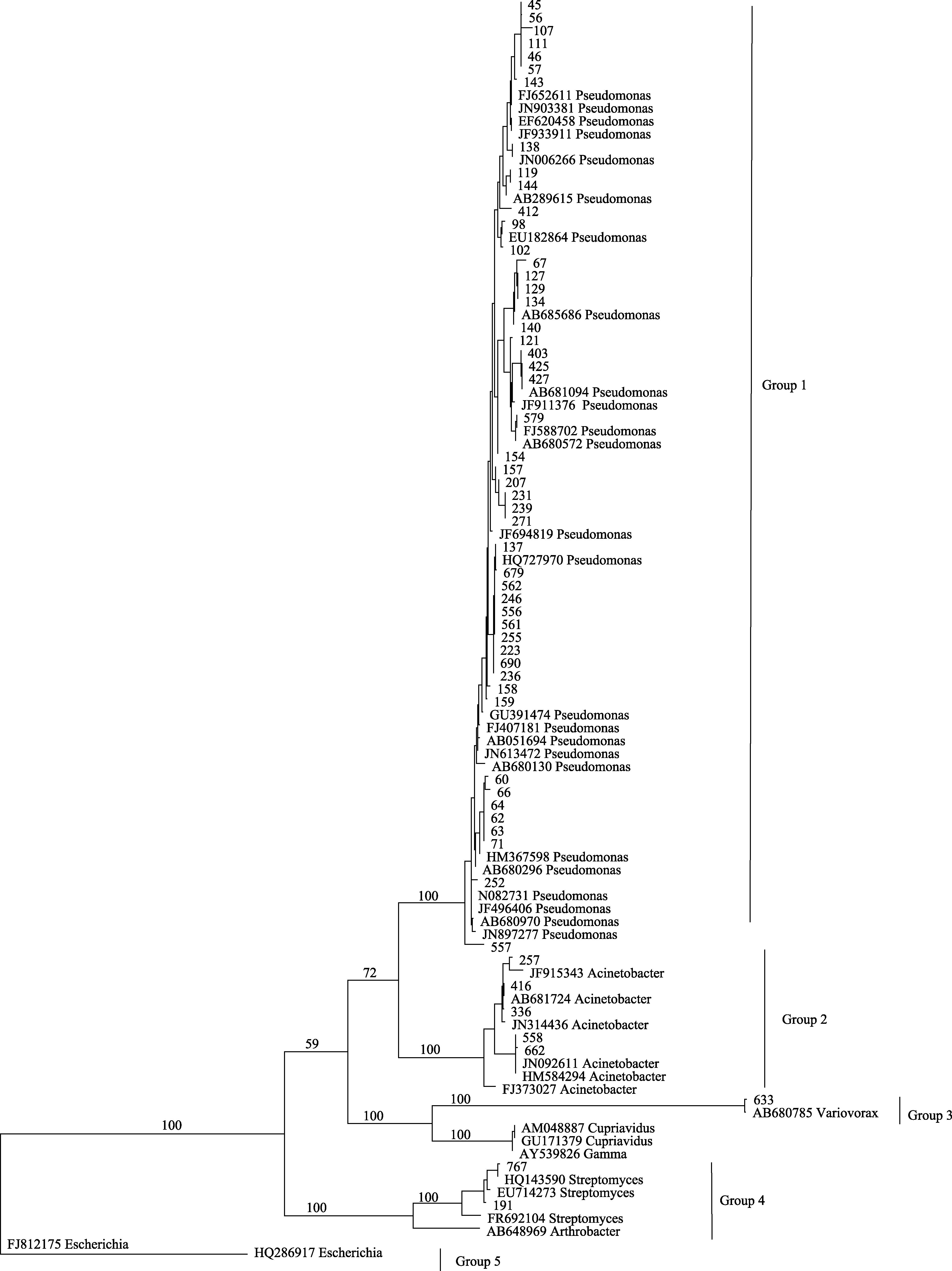
Fig. 2 One of most parsimonious trees based on the analysis of 16S rRNA gene sequences of culturable bacteria from Tuber indicum ascocarps. Numbers above branches indicate bootstrap support above 50%.
| 1 | Barbieri E, Bertini L, Rossi I, Ceccaroli P, Saltarelli R, Guidi C, Zambonelli A, Stocchi V (2005) New evidence for bacterial diversity in the ascoma of the ectomycorrhizal fungus Tuber borchii Vittad. FEMS Microbiology Letters, 247, 23-35. |
| 2 | Barbieri E, Guidi C, Bertaux J, Frey-Klett P, Garbaye J, Ceccaroli P, Saltarelli R, Zambonelli A, Stocchi V (2007) Occurrence and diversity of bacterial communities in Tuber magnatum during truffle maturation. Environmental Microbiology, 9, 2234-2246. |
| 3 | Citterio B, Cardoni P, Potenza L, Amicucci A, Stocchi V, Gola G, Nuti M (1995) >Isolation of bacteria from sporocarps of Tuber magnatum Pico, Tuber borchii Vitt. and Tuber maculatum Vitt. In: Biotechnology of Ectomycorrhizae (eds Stocchi V, Bonfante P, Nuti M), pp. 241-248. Plenum Press, New York. |
| 4 | Citterio B, Malatesta M, Battistelli S, Marcheggiani F, Baffone W, Saltarelli R, Stocchi V, Gazzanelli G (2001) Possible involvement of Pseudomonas fluorescens and Bacillaceae in structural modifications of Tuber borchii fruit bodies. Canadian Journal of Microbiology, 47, 264-268. |
| 5 | Deveau A, Antony-Babu S, Le Tacon F, Robin C, Frey-Klett P (2016) Temporal changes of bacterial communities in the Tuber melanosporum ectomycorrhizosphere during ascocarp development. Mycorrhiza, 26, 389-399. |
| 6 | Dib-Bellahouel S, Fortas Z (2014) Activity of the desert truffle Terfezia boudieri Chatin, against associated soil microflora. African Journal of Microbiology Research, 8, 3008-3016. |
| 7 | Dorofeev AG, Grigor’eva NV, Kozlov MN, Kevbrina MV, Aseeva VG, Nikolav YA (2014) Approaches to cultivation of “nonculturable” bacteria: Cyclic cultures. Microbiology, 83, 450-461. |
| 8 | Fu Y, Li X, Li Q, Wu H, Xiong C, Geng Q, Sun H, Sun Q (2016) Soil microbial communities of three major Chinese truffles in Southwest China. Canadian Journal of Microbiology, 62, 970-979. |
| 9 | Gryndler M, Soukupová L, Hršelová H, Gryndlerová H, Borovičika J, Streiblová E, Jansa J (2013) A quest for indigenous truffle helper prokaryotes. Environment Microbiology Reports, 5, 346-352. |
| 10 | Gurtler V, Stanisich VA (1996) New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology, 142, 3-16. |
| 11 | Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| 12 | Liu PG, Chen J, Deng XJ, Wang XH, Qiao P, Zhang JP, Wan SP, Geng LY, Zhao FL, Zhao WQ, Wang XJ (2016) Truffles from China. Science Press, Beijing. (in Chinese) |
| [刘培贵, 陈娟, 邓晓娟, 王向华, 乔鹏, 张介平, 万山平, 耿丽英, 赵峰岚, 赵文青, 王晓进 (2016) 中国的块菌. 科学出版社, 北京.] | |
| 13 | Mavromatis K, Land M L, Brettin T S, Quest D J, Copeland A, Clum A, Goodwin L, Woyke T, Lapidus A, Klenk HP, Cottingham RW, Kyrpides NC (2012) The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS ONE, 7, e48837. |
| 14 | Mello A, Miozzi L, Vizzini A, Napoli C, Kowalchuk G, Bonfante P (2010) Bacterial and fungal communities associated with Tuber magnatum—productive niches. Plant Biosystems, 144, 323-332. |
| 15 | Navarro-Ródenas A, Berná L M, Lozano-Carrillo C, Andrino A, Morte A (2016) Beneficial native bacteria improve survival and mycorrhization of desert truffle mycorrhizal plants in nursery conditions. Mycorrhiza, 26, 769-779. |
| 16 | Picceri GG, Leonardi P, Iotti M, Gallo M, Baldi F, Zambonelli A, Amicucci A, Vallorani L, Piccoli G, Ciccimarra G, Arshakyan M, Burattini S, Falcieri E, Chiarantini L (2018) Bacteria-produced ferric exopolysaccharide nanoparticles as iron delivery system for truffles (Tuber borchii). Applied Microbiology and Biotechnology, 102, 1429-1441. |
| 17 | Qiao P, Tian W, Liu PG, Yu GQ, Chen J, Deng XJ, Wan SP, Wang R, Wang Y, Guo HG (2018) Phylogeography and population genetic analyses reveal the speciation of the Tuber indicum complex. Fungal Genetics and Biology, 113, 14-23. |
| 18 | Sbrana C, Bagnoli G, Bedini S, Filippi C, Giovanetti M, Nuti MP (2000) Adhesion to hyphal matrix and antifungal activity of Pseudomonas strains isolated from Tuber borchii ascocarps. Canadian Journal of Microbiology, 46, 259-268. |
| 19 | Sbrana C, Agnolucci M, Bedini S, Lepera A, Toffanin A, Giovannetti M, Nuti MP (2002) Diversity of culturable bacterial populations associated to Tuber borchii ectomycorrhizas and their activity on T. borchii mycelial growth. FEMs Microbiology Letters, 211, 195-201. |
| 20 | Siqueira JF Jr, Fouad AF, Rôças IN (2012) Pyrosequencing as a tool for better understanding of human microbiomes. Journal of Oral Microbiology, 4, 10743. |
| 21 | Streiblová E, Gryndlerová H, Gryndler M (2012) Truffle brûlé: An efficient fungal life strategy. FEMS Microbiology Ecology, 80, 1-8. |
| 22 | Soudzilovskaia NA, Douma JC, Akhmetzhanova AA, Van Bodegom PM, Cornwell WK, Moens EJ, Treseder KK, Tibbett M, Wang YP, Cornelissen JHC (2015) Global patterns of plant root colonization intensity by mycorrhizal fungi explained by climate and soil chemistry. Global Ecology and Biogeography, 24, 371-382. |
| 23 | Splivallo R, Deveau A, Valdez N, Kirchhoff N, Frey-Klett P, Karlovsky P (2015) Bacteria associated with truffle fruiting bodies contribute to truffle aroma. Environmental Microbiology, 17, 2647-2660. |
| 24 | Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. |
| 25 | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| 26 | Wan SP, Liu PG (2014) Diversity of culturable bacteria associated with ascocarps of a Chinese white truffle. Plant Diversity and Resources, 36, 29-36. |
| 27 | Yan WR, Zhao TC, Xiao TB, Xiao M, Zhao ZX, Chen MC (2013) Applications of biocontrol bacterial in plant disease control. Genomics and Applied Biology, 32, 533-539. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()