



Biodiv Sci ›› 2016, Vol. 24 ›› Issue (8): 966-976. DOI: 10.17520/biods.2016057 cstr: 32101.14.biods.2016057
• Forums • Previous Articles
Received:2016-02-28
Accepted:2016-07-12
Online:2016-08-20
Published:2016-09-02
Contact:
Xie Ping
Ping Xie. How did nucleus and sexual reproduction come into being?[J]. Biodiv Sci, 2016, 24(8): 966-976.
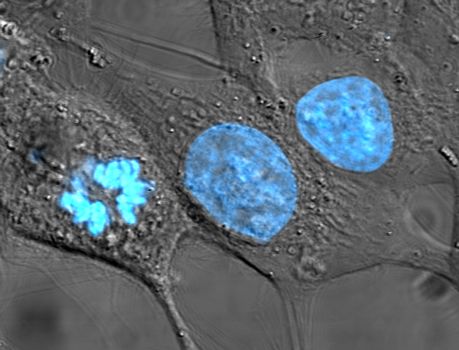
Fig. 1 Cultured HeLa cells have been stained with Hoechst turning their nuclei blue. The central and rightmost cells are in interphase, so the entire nuclei are labeled. The cell on the left is going through mitosis and its DNA has condensed (source: Wikipedia)
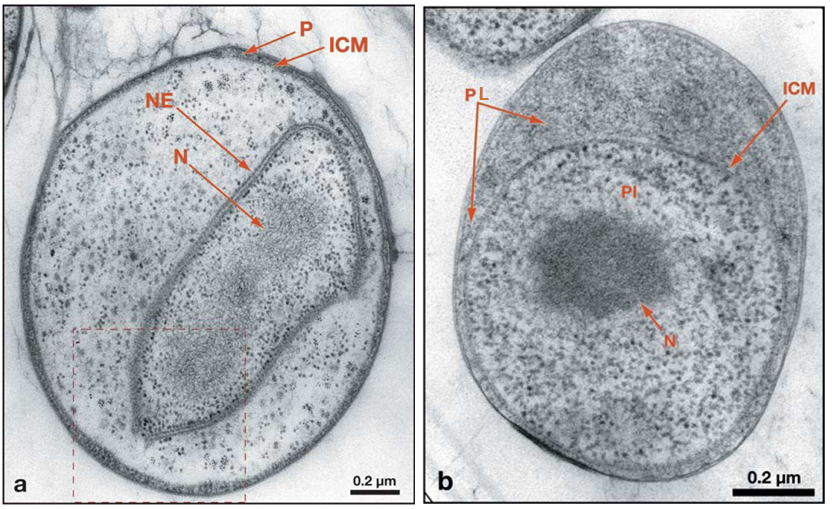
Fig. 2 (a) Transmission electron micrograph of thin section of Gemmata obscuriglobus fixed and prepared via cryosubstitution, showing the membrane-bound nuclear body with its nuclear envelope (NE) surrounding the nucleoid (N) and the more general features of the planctomycete cell plan, including the intracytoplasmic membrane (ICM) and paryphoplasm (P). (b) Transmission electron micrograph of thin section of cryosubstituted cell of Pirellula staleyi displaying compartmentalization into pirellulosome (PI) and P separated by the ICM. N is contained within the pirellulosome and thus compartmentalized and surrounded by the single ICM membrane (cited from Fuerst, 2005)
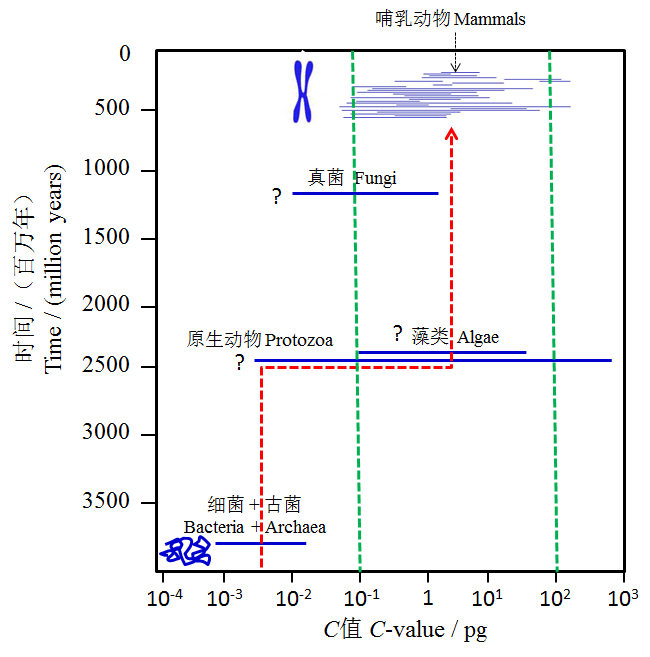
Fig. 5 Packing principle of eukaryotic DNA based on “C value” of various organisms. The dotted red line with an arrow indicates evolutionary track of the median “C value”, and the dotted green line shows the major range of the “C value” of most eukaryotes. Question marks indicate uncertainity about the dates of their origins. C values of various organisms are cited from Gregory (2004)
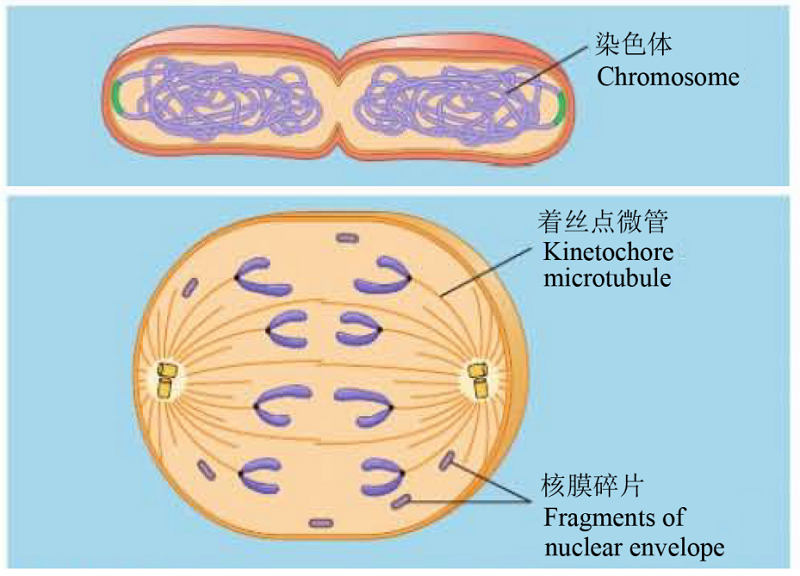
Fig. 6 Comparison of the cell divisions between bacteria (upper figure, the bacterium Escherichia coli) and eukaryotes (lower figure) (cited from Campbell & Reece, 2008)
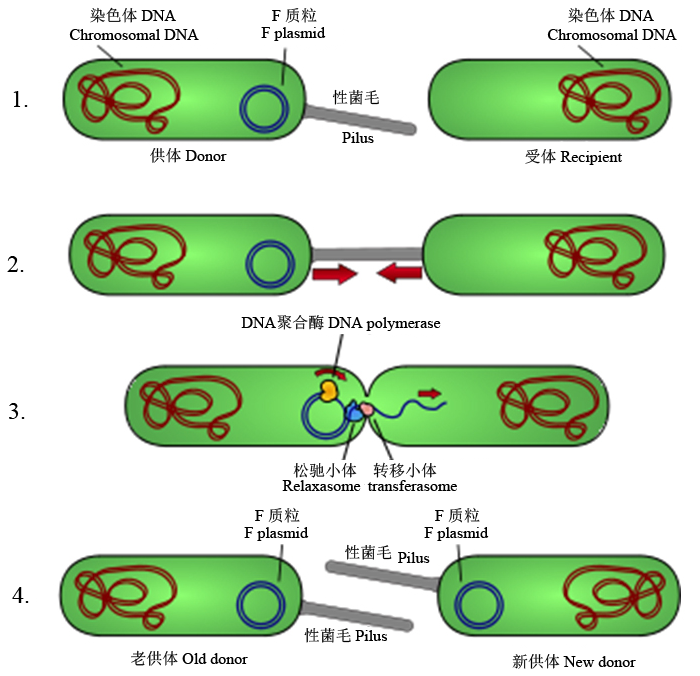
Fig. 7 Schematic drawing of bacterial conjugation. Conjugation diagram. 1, Donor cell produces pilus; 2, Pilus attaches to recipient cell and brings the two cells together; 3, The mobile plasmid is nicked and a single strand of DNA is then transferred to the recipient cell; 4, Both cells synthesize a complementary strand to produce a double stranded circular plasmid and also reproduce pili; both cells are now viable donors (cited from Wikipedia).
| [1] | Bell G (1982) The Masterpiece of Nature: the Evolution and Genetics of Sexuality. University of California Press, Berkeley. |
| [2] | Bell PJ (2001) Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? Journal of Molecular Evolution, 53, 251-256. |
| [3] | Campbell NA, Reece JB (2008) Biology, 8th edn. Benjamin Cummings, San Francisco. |
| [4] | Coyne JA (2009) Why Evolution is True. Viking Penguin, New York David LA, Alm EJ (2011) Rapid evolutionary innovation during an Archaean genetic expansion. Nature, 469, 93-96. |
| [5] | de Roos AD (2006) The origin of the eukaryotic cell based on conservation of existing interfaces. Artificial Life, 12, 513-523. |
| [6] | Ebert D (2005) Ecology, Epidemiology, and Evolution of Parasitism in Daphnia. National Library of Medicine (US), National Center for Biotechnology Information, Bethesda, MD. |
| [7] | Fuerst JA (2005) Intracellular compartmentation in planctomycetes. Annual Review of Microbiology, 59, 299-328. |
| [8] | Gregory TR (2004) Macroevolution, hierarchy theory, and the C-value enigma. Paleobiology, 30, 179-202. |
| [9] | Hadany L, Comeron JM (2008) Why are sex and recombination so common? Annals of the New York Academy of Sciences, 1133, 26-43. |
| [10] | Hogan CM (2010) Archaea. In: Encyclopedia of Earth (eds Monosson E, Cleveland C). National Council for Science and the Environment, Washington, DC. |
| [11] | Jacob F (1998) Of Flies, Mice and Men (translated from the French edition). Harvard University Press, Cambridge MA. |
| [12] | Koonin EV (2015) Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Philosophical Transactions of the Royal Society B, 370, 20140333. |
| [13] | Margulis L (1981) Symbiosis in Cell Evolution, pp. 206-227. W. H. Freeman and Company, San Francisco. |
| [14] | Martin WF, Garg S, Zimorski V (2015) Endosymbiotic theories for eukaryote origin. Philosophical Transactions of the Royal Society B, 370, 20140330. |
| [15] | McInerney J, Pisani D, O’Connell MJ (2015) The ring of life hypothesis for eukaryote origins is supported by multiple kinds of data. Philosophical Transactions of the Royal Society B, 370, 20140323 |
| [16] | Mereschkowsky K (1910) Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen. Biologisches Centralblatt 30, 353-367. |
| [17] | Moreira D, López-García P (2015) Evolution of viruses and cells: do we need a fourth domain of life to explain the origin of eukaryotes? Philosophical Transactions of the Royal Society B, 370, 20140327. |
| [18] | Pennisi E (2004) Evolutionary biology. The birth of the nucleus. Science, 305, 766-768. |
| [19] | Prescott DM (1994) The DNA of ciliated protozoa. Microbiological Reviews, 58, 233-267. |
| [20] | Raven PH, Evert RF, Eichhorn SE (1992) Biology of Plants, 5th edn. Worth Publishers, New York. |
| [21] | Schurko AM, Neiman M, Logsdon JM Jr (2008) Signs of sex: what we know and how we know it. Trends in Ecology & Evolution, 24, 208-217. |
| [22] | Takemura M (2001) Poxviruses and the origin of the eukaryotic nucleus. Journal of Molecular Evolution, 52, 419-442. |
| [23] | Villarreal L, DeFilippis V (2000) A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. Journal of Virology, 74, 7079-7084. |
| [24] | Wallin IE (1923) The mitochondria problem. The American Naturalist, 57, 255-261. |
| [25] | Xie P (2013) Scaling Ecology to Understand Natural Design of Life Systems and Their Operations and Evolutions—Integration of Ecology, Genetics and Evolution Through Reproduction. Science Press, Beijing. |
| [谢平 (2013) 从生态学透视生命系统的设计、运作与演化——生态、遗传和进化通过生殖的融合. 科学出版社, 北京.] | |
| [26] | Xie P (2014) The Aufhebung and Breakthrough of the Theories on the Origin and Evolution of Life—Life in Philosophy and Philosophy in Life Sciences. Science Press, Beijing. |
| [谢平 (2014) 生命的起源-进化理论之扬弃与革新——哲学中的生命, 生命中的哲学. 科学出版社, 北京.] | |
| [27] | Zimmer C (2009) On the origin of sexual reproduction. Science, 324, 1254-1256. |
| [1] | Jiabei He, Ke Ke, Haiming Sun, Liping Hu, Xiaowei Zhao, Wenhao Wang, Qiang Zhao. Diet analysis of Neptunea cumingii using metabarcoding [J]. Biodiv Sci, 2025, 33(1): 24403-. |
| [2] | Yihui Jiang, Yue Liu, Xu Zeng, Zheying Lin, Nan Wang, Jihao Peng, Ling Cao, Cong Zeng. Fish diversity and connectivity in six national marine protected areas in the East China Sea [J]. Biodiv Sci, 2024, 32(6): 24128-. |
| [3] | Weijie Shu, Hua He, Luo Zeng, Zhirong Gu, Dunyan Tan, Xiaochen Yang. Spatial distribution and sexual dimorphism of dioecious Arisaema erubescens [J]. Biodiv Sci, 2024, 32(6): 24084-. |
| [4] | Lejie Wu, Zekang Liu, Xing Tian, Qun Zhang, Bo Li, Jihua Wu. Effects of genotypic diversity on vegetative growth and reproductive strategies of Scirpus mariqueter population [J]. Biodiv Sci, 2024, 32(4): 23478-. |
| [5] | Xiaoyan Luo, Qiang Li, Xiaolei Huang. DNA barcode reference dataset for flower-visiting insects in Daiyun Mountain National Nature Reserve [J]. Biodiv Sci, 2023, 31(8): 23236-. |
| [6] | Chao Xing, Yi Lin, Zhiqiang Zhou, Lianjun Zhao, Shiwei Jiang, Zhenzhen Lin, Jiliang Xu, Xiangjiang Zhan. The establishment of terrestrial vertebrate genetic resource bank and species identification based on DNA barcoding in Wanglang National Nature Reserve [J]. Biodiv Sci, 2023, 31(7): 22661-. |
| [7] | Zhiyuan Dong, Linlin Chen, Naipeng Zhang, Li Chen, Debin Sun, Yanmei Ni, Baoquan Li. Response of fish diversity to hydrological connectivity of typical tidal creek system in the Yellow River Delta based on environmental DNA metabarcoding [J]. Biodiv Sci, 2023, 31(7): 23073-. |
| [8] | Zhenjie Zhan, Chao Zhang, Minhao Chen, Jiadong Wang, Aihua Fu, Yuwei Fan, Xiaofeng Luan. DNA metabarcoding-based winter diet analysis of Eurasian otter (Lutra lutra) in the northern Greater Khingan Mountains [J]. Biodiv Sci, 2023, 31(6): 22586-. |
| [9] | Fan Wu, Shenyun Liu, Huqiang Jiang, Qian Wang, Kaiwei Chen, Hongliang Li. Pollination difference between Apis cerana cerana and Apis mellifera ligustica during the late autumn and winter [J]. Biodiv Sci, 2023, 31(5): 22528-. |
| [10] | Buqing Peng, Ling Tao, Jing Li, Ronghui Fan, Shunde Chen, Changkun Fu, Qiong Wang, Keyi Tang. DNA metabarcoding dietary analysis of six sympatric small mammals at the Laojunshan National Nature Reserve, Sichuan Province [J]. Biodiv Sci, 2023, 31(4): 22474-. |
| [11] | Mei Shen, Ningning Guo, Zunlan Luo, Xiaochen Guo, Guang Sun, Nengwen Xiao. Explore the distribution and influencing factors of fish in major rivers in Beijing with eDNA metabarcoding technology [J]. Biodiv Sci, 2022, 30(7): 22240-. |
| [12] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [13] | Dan Peng, Zhiqiang Wu. Progress on sex determination of dioecious plants [J]. Biodiv Sci, 2022, 30(3): 21416-. |
| [14] | Yixin Sun, Yingbin Li, Yuhui Li, Bing Li, Xiaofang Du, Qi Li. Application of high-throughput sequencing technique in the study of nematode diversity [J]. Biodiv Sci, 2022, 30(12): 22266-. |
| [15] | Shanlin Liu, Na Qiu, Shuyi Zhang, Zhunan Zhao, Xin Zhou. Application of genomics technology in biodiversity conservation research [J]. Biodiv Sci, 2022, 30(10): 22441-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()
