



Biodiv Sci ›› 2023, Vol. 31 ›› Issue (6): 22586. DOI: 10.17520/biods.2022586 cstr: 32101.14.biods.2022586
• Original Papers: Animal Diversity • Previous Articles Next Articles
Zhenjie Zhan1, Chao Zhang2, Minhao Chen1, Jiadong Wang1, Aihua Fu1, Yuwei Fan1, Xiaofeng Luan1,*( )
)
Received:2022-10-18
Accepted:2023-02-05
Online:2023-06-20
Published:2023-04-18
Contact:
* E-mail: luanxiaofeng@bjfu.edu.cn
Zhenjie Zhan, Chao Zhang, Minhao Chen, Jiadong Wang, Aihua Fu, Yuwei Fan, Xiaofeng Luan. DNA metabarcoding-based winter diet analysis of Eurasian otter (Lutra lutra) in the northern Greater Khingan Mountains[J]. Biodiv Sci, 2023, 31(6): 22586.
| 引物 Primer | 扩增区域 Target | 引物序列 Primer sequence (5'-3') | 产物长度 Product length | 参考文献 Reference |
|---|---|---|---|---|
| LLCBL/R | Cytb | F: AAAGCCACCCTGACACGATT | 80 bp | Thomsen et al, |
| R: AGCAGGTGGATTGTTGCTAGTG | ||||
| 12S V5 | 12S rRNA | F: TAGAACAGGCTCCTCTAG | 98 bp | Riaz et al, |
| R: TTAGATACCCCACTATGC | ||||
| OBS1 | - | CTATGCTCAGCCCTAAACATAGATAGCTTACATAACAAAACTATCTGC-C3 | - | Kumari et al, |
| HomoB | - | CTATGCTTAGCCCTAAACCTCAACAGTTAAATCAACAAAACTGCT-C3 | - | De Barba et al, |
| BF2/BR1 | COI | F: GCHCCHGAYATRGCHTTYCC | 322 bp | Elbrecht & Leese, |
| R: ARYATDGTRATDGCHCCDGC |
Table 1 Sequences of the primers used in the study
| 引物 Primer | 扩增区域 Target | 引物序列 Primer sequence (5'-3') | 产物长度 Product length | 参考文献 Reference |
|---|---|---|---|---|
| LLCBL/R | Cytb | F: AAAGCCACCCTGACACGATT | 80 bp | Thomsen et al, |
| R: AGCAGGTGGATTGTTGCTAGTG | ||||
| 12S V5 | 12S rRNA | F: TAGAACAGGCTCCTCTAG | 98 bp | Riaz et al, |
| R: TTAGATACCCCACTATGC | ||||
| OBS1 | - | CTATGCTCAGCCCTAAACATAGATAGCTTACATAACAAAACTATCTGC-C3 | - | Kumari et al, |
| HomoB | - | CTATGCTTAGCCCTAAACCTCAACAGTTAAATCAACAAAACTGCT-C3 | - | De Barba et al, |
| BF2/BR1 | COI | F: GCHCCHGAYATRGCHTTYCC | 322 bp | Elbrecht & Leese, |
| R: ARYATDGTRATDGCHCCDGC |
| 物种类别 Species taxa | 出现样品数 Occurrence | 相对出现频率 %RFO | 相对序列丰度 %RRA | 生物类群 Biome |
|---|---|---|---|---|
| 杂色杜父鱼 Cottus poecilopus | 30 | 19.35 | 27.32 | 鱼类 Fish |
| 黑龙江林蛙 Rana amurensis | 24 | 15.48 | 21.73 | 蛙类 Frog |
| 克氏杜父鱼 Cottus czerskii | 15 | 9.68 | 16.47 | 鱼类 Fish |
| 东北林蛙 Rana dybowskii | 4 | 2.58 | 7.21 | 蛙类 Frog |
| 须鳅属一种 Barbatula sp. | 13 | 8.39 | 5.47 | 鱼类 Fish |
| 真鱥 Phoxinus phoxinus | 18 | 11.61 | 5.09 | 鱼类 Fish |
| 湖大吻鱥 Rhynchocypris percnurus | 10 | 6.45 | 4.65 | 鱼类 Fish |
| 黑龙江茴鱼 Thymallus arcticus | 8 | 5.16 | 2.96 | 鱼类 Fish |
| 拉氏大吻鱥 Rhynchocypris lagowskii | 8 | 5.16 | 2.81 | 鱼类 Fish |
| 黑龙江中杜父鱼 Mesocottus haitej | 2 | 1.29 | 1.54 | 鱼类 Fish |
| 茴鱼属一种 Thymallus sp. | 4 | 2.58 | 1.43 | 鱼类 Fish |
| 七鳃鳗属一种 Lampetra sp. | 4 | 2.58 | 1.08 | 鱼类 Fish |
| 葛氏鲈塘鳢 Perccottus glenii | 5 | 3.23 | 0.81 | 鱼类 Fish |
| 江鳕 Lota lota | 4 | 2.58 | 0.66 | 鱼类 Fish |
| 杜父鱼属一种 Cottus sp. | 3 | 1.94 | 0.39 | 鱼类 Fish |
| 大吻鱥属一种 Rhynchocypris sp. | 2 | 1.29 | 0.33 | 鱼类 Fish |
| 花鳅属一种 Cobitis sp. | 1 | 0.65 | 0.05 | 鱼类 Fish |
Table 2 Results of diet analysis in 12S rRNA region (N = 35) of Eurasian otter fecal samples in the northern Greater Khingan Mountains. %RFO, Relative frequency of occurrence; %RRA, Relative read abundance.
| 物种类别 Species taxa | 出现样品数 Occurrence | 相对出现频率 %RFO | 相对序列丰度 %RRA | 生物类群 Biome |
|---|---|---|---|---|
| 杂色杜父鱼 Cottus poecilopus | 30 | 19.35 | 27.32 | 鱼类 Fish |
| 黑龙江林蛙 Rana amurensis | 24 | 15.48 | 21.73 | 蛙类 Frog |
| 克氏杜父鱼 Cottus czerskii | 15 | 9.68 | 16.47 | 鱼类 Fish |
| 东北林蛙 Rana dybowskii | 4 | 2.58 | 7.21 | 蛙类 Frog |
| 须鳅属一种 Barbatula sp. | 13 | 8.39 | 5.47 | 鱼类 Fish |
| 真鱥 Phoxinus phoxinus | 18 | 11.61 | 5.09 | 鱼类 Fish |
| 湖大吻鱥 Rhynchocypris percnurus | 10 | 6.45 | 4.65 | 鱼类 Fish |
| 黑龙江茴鱼 Thymallus arcticus | 8 | 5.16 | 2.96 | 鱼类 Fish |
| 拉氏大吻鱥 Rhynchocypris lagowskii | 8 | 5.16 | 2.81 | 鱼类 Fish |
| 黑龙江中杜父鱼 Mesocottus haitej | 2 | 1.29 | 1.54 | 鱼类 Fish |
| 茴鱼属一种 Thymallus sp. | 4 | 2.58 | 1.43 | 鱼类 Fish |
| 七鳃鳗属一种 Lampetra sp. | 4 | 2.58 | 1.08 | 鱼类 Fish |
| 葛氏鲈塘鳢 Perccottus glenii | 5 | 3.23 | 0.81 | 鱼类 Fish |
| 江鳕 Lota lota | 4 | 2.58 | 0.66 | 鱼类 Fish |
| 杜父鱼属一种 Cottus sp. | 3 | 1.94 | 0.39 | 鱼类 Fish |
| 大吻鱥属一种 Rhynchocypris sp. | 2 | 1.29 | 0.33 | 鱼类 Fish |
| 花鳅属一种 Cobitis sp. | 1 | 0.65 | 0.05 | 鱼类 Fish |
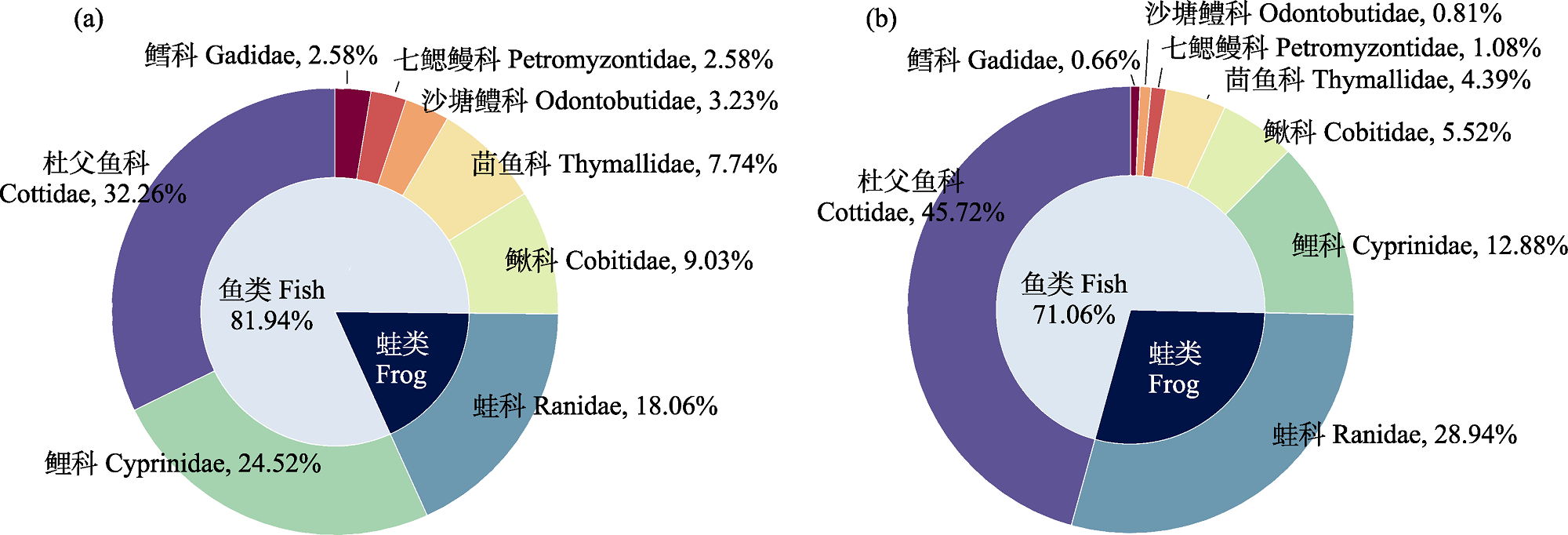
Fig. 2 Vertebrate of Eurasian otter feces (N = 35) in northern Greater Khingan Mountains at family level. (a) Relative frequency of occurrence; (b) Relative read abundance.
| 物种类别 Species taxa | 出现样品数 Occurrence | 相对出现频率 %RFO | 相对序列丰度 %RRA | 生物类群 Biome |
|---|---|---|---|---|
| 蜻蜓目 Odonata | 8 | 20.51 | 33.86 | 昆虫 Insect |
| 毛翅目 Trichoptera | 9 | 23.08 | 25.92 | 昆虫 Insect |
| 襀翅目 Plecoptera | 7 | 17.95 | 18.74 | 昆虫 Insect |
| 蜉蝣目 Ephemeroptera | 8 | 20.51 | 15.87 | 昆虫 Insect |
| 双翅目 Diptera | 7 | 17.95 | 5.60 | 昆虫 Insect |
Table 3 Results of diet analysis in COI region (N = 12) of Eurasian otter fecal samples in the northern Greater Khingan Mountains. %RFO, Relative frequency of occurrence; %RRA, Relative read abundance.
| 物种类别 Species taxa | 出现样品数 Occurrence | 相对出现频率 %RFO | 相对序列丰度 %RRA | 生物类群 Biome |
|---|---|---|---|---|
| 蜻蜓目 Odonata | 8 | 20.51 | 33.86 | 昆虫 Insect |
| 毛翅目 Trichoptera | 9 | 23.08 | 25.92 | 昆虫 Insect |
| 襀翅目 Plecoptera | 7 | 17.95 | 18.74 | 昆虫 Insect |
| 蜉蝣目 Ephemeroptera | 8 | 20.51 | 15.87 | 昆虫 Insect |
| 双翅目 Diptera | 7 | 17.95 | 5.60 | 昆虫 Insect |
| [1] |
Alberdi A, Aizpurua O, Bohmann K, Gopalakrishnan S, Lynggaard C, Nielsen M, Gilbert MTP (2019) Promises and pitfalls of using high-throughput sequencing for diet analysis. Molecular Ecology Resources, 19, 327-348.
DOI PMID |
| [2] | Bokulich NA, Dillon MR, Bolyen E, Kaehler BD, Huttley GA, Caporaso JG (2018) q2-sample-classifier: Machine-learning tools for microbiome classification and regression. Journal of Open Research Software, 3, 934. |
| [3] | Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo JR, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang LJ, Kaehler BD, Bin KK, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Lai Preuss M, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan YH, Wang MX, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang YL, Zhu QY, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37, 852-857. |
| [4] | Britton R, Pegg J, Shepherd JS (2006) Revealing the prey items of the otter Lutra lutra in South West England using stomach contents analysis. Folia Zoologica, 55, 167-174. |
| [5] |
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13, 581-583.
DOI PMID |
| [6] |
Carss DN, Parkinson SG (1996) Errors associated with otter Lutra lutra faecal analysis. I. Assessing general diet from spraints. Journal of Zoology, 238, 301-317.
DOI URL |
| [7] |
Clavero M, Prenda J, Delibes M (2003) Trophic diversity of the otter (Lutra lutra L.) in temperate and Mediterranean freshwater habitats. Journal of Biogeography, 30, 761-769.
DOI URL |
| [8] |
De Barba M, Miquel C, Boyer F, Mercier C, Rioux D, Coissac E, Taberlet P (2014) DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet. Molecular Ecology Resources, 14, 306-323.
DOI PMID |
| [9] |
Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, Kartzinel TR, Eveson JP (2019) Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Molecular Ecology, 28, 391-406.
DOI PMID |
| [10] | Duan LL (2017) Impacts of Changes in Climate, Forest Cover and Permafrost on Streamflow in the Da Hinggan Mountains watersheds. PhD dissertation, Northeast Forestry University, Harbin. (in Chinese with English abstract) |
| [段亮亮 (2017) 大兴安岭气候、森林覆盖率和冻土变化对河川径流的影响. 博士学位论文, 东北林业大学, 哈尔滨.] | |
| [11] |
Duffy JE, Cardinale BJ, France KE, McIntyre PB, Thébault E, Loreau M (2007) The functional role of biodiversity in ecosystems: Incorporating trophic complexity. Ecology Letters, 10, 522-538.
PMID |
| [12] | Elbrecht V, Leese F (2017) Validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment. Frontiers in Environmental Science, 5, 11. |
| [13] |
Erlinge S (1967) Home range of the otter Lutra lutra L. in southern Sweden. Oikos, 18, 186-209.
DOI URL |
| [14] | Gomez L, Leupen B, Theng M, Fernandez K, Savage M (2016) Illegal Otter Trade: An Analysis of Seizures in Selected Asian Countries (1980-2015). Traffic, Malaysia. |
| [15] |
Han XS, Dong ZY, Zhao G, Zhao X, Shi XY, Lü Z, Li HQ (2021) Using surveillance cameras to analyze the activity pattern of the Eurasian otters (Lutra lutra) and the efficiency of camera trap monitoring. Biodiversity Science, 29, 770-779. (in Chinese with English abstract)
DOI URL |
| [韩雪松, 董正一, 赵格, 赵翔, 史湘莹, 吕植, 李宏奇 (2021) 基于视频监控系统的欧亚水獭活动节律初报及红外相机监测效果评估. 生物多样性, 29, 770-779.] | |
| [16] |
Harper L, Watson H, Donnelly R, Hampshire R, Sayer C, Breithaupt T, Hänfling B (2020) Using DNA metabarcoding to investigate diet and niche partitioning in the native European otter (Lutra lutra) and invasive American mink (Neovison vison). Metabarcoding and Metagenomics, 4, e56087.
DOI URL |
| [17] |
Hong SW, Gim JS, Kim HG, Cowan PE, Joo GJ (2019) A molecular approach to identifying the relationship between resource use and availability in Eurasian otters (Lutra lutra). Canadian Journal of Zoology, 97, 797-804.
DOI URL |
| [18] | Huo TB, Song XS, Wang ZY, Wang Y, Li TZ (2015) Biological characteristics of alpine sculpin Cottus poecilopus Heckel. Chinese Journal of Fisheries, 28, 16-20. (in Chinese with English abstract) |
| [霍堂斌, 宋雪松, 王振洋, 王野, 栗铁柱 (2015) 杂色杜父鱼生物学初步研究. 水产学杂志, 28, 16-20.] | |
| [19] | Kartzinel TR, Chen PA, Coverdale TC, Erickson DL, Kress WJ, Kuzmina ML, Rubenstein DI, Wang W, Pringle RM (2015) DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proceedings of the National Academy of Sciences, USA, 112, 8019-8024. |
| [20] |
Kelly RP, Port JA, Yamahara KM, Crowder LB (2014) Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE, 9, e86175.
DOI URL |
| [21] |
Klare U, Kamler JF, Macdonald DW (2011) A comparison and critique of different scat-analysis methods for determining carnivore diet. Mammal Review, 41, 294-312.
DOI URL |
| [22] |
Krawczyk AJ, Bogdziewicz M, Majkowska K, Glazaczow A (2016) Diet composition of the Eurasian otter Lutra lutra in different freshwater habitats of temperate Europe: A review and meta-analysis. Mammal Review, 46, 106-113.
DOI URL |
| [23] | Kruuk H (2006) Otters:Ecology, Behaviour and Conservation. Oxford University Press, New York. |
| [24] | Kumari P, Dong K, Eo KY, Lee WS, Kimura J, Yamamoto N (2019) DNA metabarcoding-based diet survey for the Eurasian otter (Lutra lutra): Development of a Eurasian otter-specific blocking oligonucleotide for 12S rRNA gene sequencing for vertebrates PLoS ONE, 14, e0226253. |
| [25] |
Lanszki J, Lehoczky I, Kotze A, Somers MJ (2016) Diet of otters (Lutra lutra) in various habitat types in the Pannonian biogeographical region compared to other regions of Europe. PeerJ, 4, e2266.
DOI URL |
| [26] |
Li F, Chan BPL (2018) Past and present: The status and distribution of otters (Carnivora: Lutrinae) in China. Oryx, 52, 619-626.
DOI URL |
| [27] | Lin FH, Sun CM, Chen WL, Wang JG (2016) Community structure and bioindicator of benthic fauna from Huma to Heihe in the upper reaches of Heilongjiang Province. Journal of Mudanjiang Normal University (Natural Sciences Edition), 22, 57-59. (in Chinese) |
| [林繁会, 孙春梅, 陈伟莉, 王金刚 (2016) 黑龙江上游呼玛至黑河江段底栖动物群落结构及其生物指示作用. 牡丹江师范学院学报(自然科学版), 22, 57-59.] | |
| [28] | Lü J, Yang L, Yang L, Li JX, Huang MJ, Luan XF (2018) Potential distribution of otter in Northeast China. Journal of Fujian Agriculture and Forestry University (Natural Science Edition), 47, 473-479. (in Chinese with English abstract) |
| [吕江, 杨立, 杨蕾, 李婧昕, 黄木娇, 栾晓峰 (2018) 中国东北地区水獭种群潜在分布区的预测. 福建农林大学学报(自然科学版), 47, 473-479.] | |
| [29] |
Lu Q, Hu Q, Shi XG, Jin SL, Li S, Yao M (2019) Metabarcoding diet analysis of snow leopards (Panthera uncia) in Wolong National Nature Reserve, Sichuan Province. Biodiversity Science, 27, 960-969. (in Chinese with English abstract)
DOI |
|
[陆琪, 胡强, 施小刚, 金森龙, 李晟, 姚蒙 (2019) 基于分子宏条形码分析四川卧龙国家级自然保护区雪豹的食性. 生物多样性, 27, 960-969.]
DOI |
|
| [30] | Marcolin F, Iordan F, Pizzul E, Pallavicini A, Torboli V, Manfrin C, Quaglietta L (2020) Otter diet and prey selection in a recently recolonized area assessed using microscope analysis and DNA barcoding. Hystrix, 31, 64-72. |
| [31] |
Monterroso P, Godinho R, Oliveira T, Ferreras P, Kelly M, Morin D, Waits L, Alves P, Mills L (2019) Feeding ecological knowledge: The underutilised power of faecal DNA approaches for carnivore diet analysis. Mammal Review, 49, 97-112.
DOI |
| [32] |
Morin DJ, Higdon SD, Holub JL, Montague DM, Fies ML, Waits LP, Kelly MJ (2016) Bias in carnivore diet analysis resulting from misclassification of predator scats based on field identification. Wildlife Society Bulletin, 40, 669-677.
DOI URL |
| [33] |
Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L (2004) Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Molecular Ecology, 13, 2089-2094.
DOI PMID |
| [34] |
Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SN, Taberlet P (2012) Who is eating what: Diet assessment using next generation sequencing. Molecular Ecology, 21, 1931-1950.
DOI PMID |
| [35] | Ren ML (1981) Heilongjiang River Fish. Heilongjiang People’s Publishing House, Harbin. (in Chinese) |
| [任慕莲 (1981) 黑龙江鱼类. 黑龙江人民出版社, 哈尔滨.] | |
| [36] |
Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E (2011) ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Research, 39, e145.
DOI URL |
| [37] |
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, e2584.
DOI URL |
| [38] | Ruiz-Olmo J, Calvo A, Palazón S, Arqued V (1998) Is the otter a bioindicator? Galemys, 10, 227-237. |
| [39] |
Schmiedová L, Tomášek O, Pinkasová H, Albrecht T, Kreisinger J (2022) Variation in diet composition and its relation to gut microbiota in a passerine bird. Scientific Reports, 12, 3787.
DOI PMID |
| [40] |
Severud WJ, Windels SK, Belant JL, Bruggink JG (2013) The role of forage availability on diet choice and body condition in American beavers (Castor canadensis). Mammalian Biology, 78, 87-93.
DOI URL |
| [41] |
Shao XN, Lu Q, Liu MZ, Xiong MY, Bu HL, Wang DJ, Liu SY, Zhao JD, Li S, Yao M (2021) Generalist carnivores can be effective biodiversity samplers of terrestrial vertebrates. Frontiers in Ecology and the Environment, 19, 557-563.
DOI URL |
| [42] |
Shao XN, Song DZ, Huang QW, Li S, Yao M (2019) Fast surveys and molecular diet analysis of carnivores based on fecal DNA and metabarcoding. Biodiversity Science, 27, 543-556. (in Chinese with English abstract)
DOI |
|
[邵昕宁, 宋大昭, 黄巧雯, 李晟, 姚蒙 (2019) 基于粪便DNA及宏条形码技术的食肉动物快速调查及食性分析. 生物多样性, 27, 543-556.]
DOI |
|
| [43] |
Sheppard SK, Harwood JD (2005) Advances in molecular ecology: Tracking trophic links through predator-prey food-webs. Functional Ecology, 19, 751-762.
DOI URL |
| [44] | Shi GQ, Guo YS, Luo YM, Gong ZC, Zhang GL, Piao ZJ, Wang ZC (2021) Spring ashore activity rhythm of Eurasian otters (Lutra lutra) in Changbai Mountain, Jilin Province. Chinese Journal of Wildlife, 42, 693-699. (in Chinese with English abstract) |
| [史国强, 郭艳双, 罗玉梅, 巩振财, 张国利, 朴正吉, 王卓聪 (2021) 吉林长白山地区水獭春季岸上活动节律. 野生动物学报, 42, 693-699.] | |
| [45] | Smiroldo G, Balestrieri A, Remonti L, Prigioni C (2009) Seasonal and habitat-related variation of otter Lutra lutra diet in a Mediterranean river catchment (Italy). Folia Zoologica, 58, 87-97. |
| [46] |
Smiroldo G, Villa A, Tremolada P, Gariano P, Balestrieri A, Delfino M (2019) Amphibians in Eurasian otter Lutra lutra diet: Osteological identification unveils hidden prey richness and male-biased predation on anurans. Mammal Review, 49, 240-255.
DOI |
| [47] | Spaulding R, Krausman P, Ballard W (2000) Observer bias and analysis of gray wolf diets from scats. Wildlife Society Bulletin, 28, 947-950. |
| [48] | Taastrøm HM, Jacobsen L (1999) The diet of otters (Lutra lutra L.) in Danish freshwater habitats: Comparisons of prey fish populations. Journal of Zoology, 248, 1-13. |
| [49] |
Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Molecular Ecology, 21, 2045-2050.
DOI PMID |
| [50] | Tang FJ, Jiang ZF, Dong CZ, Ma B, Zhan PR (2007) The Zoobenthos species in Huma River. Chinese Journal of Fisheries, 20, 76-78. (in Chinese with English abstract) |
| [唐富江, 姜作发, 董崇智, 马波, 战培荣 (2007) 呼玛河底栖动物种类组成. 水产学杂志, 20, 76-78.] | |
| [51] | The Biodiversity Committee of Chinese Academy of Sciences (2022) Catalogue of Life China:2022 Annual Checklist. http://www.sp2000.org.cn. (accessed on 2022-10-07) |
| [52] |
Thomsen PF, Kielgast J, Iversen LL, Wiuf C, Rasmussen M, Gilbert MT, Orlando L, Willerslev E (2012) Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology, 21, 2565-2573.
DOI PMID |
| [53] |
Traugott M, Thalinger B, Wallinger C, Sint D (2020) Fish as predators and prey: DNA-based assessment of their role in food webs. Journal of Fish Biology, 98, 367-382.
DOI URL |
| [54] |
Trites AW, Joy R (2005) Dietary analysis from fecal samples: How many scats are enough? Journal of Mammalogy, 86, 704-712.
DOI URL |
| [55] |
Wang QY, Wang ZC, Zheng KD, Zhang P, Shen LM, Chen WL, Fan PF, Zhang L (2022) Assessing the diet of a predator using a DNA metabarcoding approach. Frontiers in Ecology and Evolution, 10, 902412.
DOI URL |
| [56] | Wang QY, Zheng KD, Han XS, He F, Zhao X, Fan PF, Zhang L (2021) Site-specific and seasonal variation in habitat use of Eurasian otters (Lutra lutra) in Western China: Implications for conservation. Zoological Research, 42, 824-832. |
| [57] |
Wasser SK, Houston CS, Koehler GM, Cadd GG, Fain SR (1997) Techniques for application of faecal DNA methods to field studies of Ursids. Molecular Ecology, 6, 1091-1097.
DOI PMID |
| [58] |
Weber JM (1990) Seasonal exploitation of amphibians by otters (Lutra lutra) in north-east Scotland. Journal of Zoology, 220, 641-651.
DOI URL |
| [59] |
Xiong MY, Wang DJ, Bu HL, Shao XN, Zhang D, Li S, Wang RJ, Yao M (2017) Molecular dietary analysis of two sympatric felids in the Mountains of Southwest China biodiversity hotspot and conservation implications. Scientific Reports, 7, 41909.
DOI PMID |
| [60] | Xu HL (1984) Species of otters in China and the conservation of their natural resources. Chinese Journal of Wildlife, 1, 9-11. (in Chinese with English abstract) |
| [徐龙辉 (1984) 中国水獭种类及资源保护. 野生动物, 1, 9-11.] | |
| [61] | Xu W (2013) Studies on Molecular Phylogeny of Rhynchocypris Fishes (Teleiostei: Cyprinidae) and Phylogeography of Cold-adapting Freshwater Fishes in the Eastern China. PhD dissertation, Fudan University, Shanghai. (in Chinese with English abstract) |
| [许旺 (2013) 鲤科大吻鱥属鱼类的分子系统发育关系与我国东部地区冷水性淡水鱼类的亲缘生物地理. 博士学位论文, 复旦大学, 上海.] | |
| [62] | Yang FY (1998) Fish resources in the northern Greater Khingan Mountains. Territory & Natural Resources Study, 20, 49-53. (in Chinese) |
| [杨富亿 (1998) 大兴安岭北部鱼类资源现状. 国土与自然资源研究, 20, 49-53.] | |
| [63] |
Zhang C, Chen MH, Yang L, Zhuang HF, Wu SH, Zhan ZJ, Wang JD, Luan XF (2022) Distribution pattern and identification of conservation priority areas of the otter in Northeast China. Biodiversity Science, 30, 21157. (in Chinese with English abstract)
DOI |
|
[张超, 陈敏豪, 杨立, 庄鸿飞, 武曙红, 湛振杰, 王嘉栋, 栾晓峰 (2022) 东北地区水獭分布格局与保护优先区识别. 生物多样性, 30, 21157.]
DOI |
|
| [64] |
Zhang L, Wang QY, Yang L, Li F, Chan BPL, Xiao ZS, Li S, Song DZ, Piao ZJ, Fan PF (2018) The neglected otters in China: Distribution change in the past 400 years and current conservation status. Biological Conservation, 228, 259-267.
DOI URL |
| [65] | Zhao WG, Ma B, Gao ZS, Liu P, Peng YL, Yu D, Liu ZT, Li L, Chen H (2021) Manual of Fish Identification in Heilongjiang River Basin (China). Science Press, Beijing. (in Chinese) |
| [赵文阁, 马波, 高智晟, 刘鹏, 彭一良, 于东, 刘志涛, 李雷, 陈辉 (2021) 黑龙江流域(中国)鱼类识别手册. 科学出版社, 北京.] |
| [1] | Jiabei He, Ke Ke, Haiming Sun, Liping Hu, Xiaowei Zhao, Wenhao Wang, Qiang Zhao. Diet analysis of Neptunea cumingii using metabarcoding [J]. Biodiv Sci, 2025, 33(1): 24403-. |
| [2] | Zhiyuan Dong, Linlin Chen, Naipeng Zhang, Li Chen, Debin Sun, Yanmei Ni, Baoquan Li. Response of fish diversity to hydrological connectivity of typical tidal creek system in the Yellow River Delta based on environmental DNA metabarcoding [J]. Biodiv Sci, 2023, 31(7): 23073-. |
| [3] | Buqing Peng, Ling Tao, Jing Li, Ronghui Fan, Shunde Chen, Changkun Fu, Qiong Wang, Keyi Tang. DNA metabarcoding dietary analysis of six sympatric small mammals at the Laojunshan National Nature Reserve, Sichuan Province [J]. Biodiv Sci, 2023, 31(4): 22474-. |
| [4] | Mei Shen, Ningning Guo, Zunlan Luo, Xiaochen Guo, Guang Sun, Nengwen Xiao. Explore the distribution and influencing factors of fish in major rivers in Beijing with eDNA metabarcoding technology [J]. Biodiv Sci, 2022, 30(7): 22240-. |
| [5] | Xuesong Han, Zhengyi Dong, Ge Zhao, Xiang Zhao, Xiangying Shi, Zhi Lü, Hongqi Li. Using surveillance cameras to analyze the activity pattern of the Eurasian otters (Lutra lutra) and the efficiency of camera trap monitoring [J]. Biodiv Sci, 2021, 29(6): 770-779. |
| [6] | Qi Lu,Qiang Hu,Xiaogang Shi,Senlong Jin,Sheng Li,Meng Yao. Metabarcoding diet analysis of snow leopards (Panthera uncia) in Wolong National Nature Reserve, Sichuan Province [J]. Biodiv Sci, 2019, 27(9): 960-969. |
| [7] | Shao Xinning, Song Dazhao, Huang Qiaowen, Li Sheng, Yao Meng. Fast surveys and molecular diet analysis of carnivores based on fecal DNA and metabarcoding [J]. Biodiv Sci, 2019, 27(5): 543-556. |
| [8] | Li Hanxi, Huang Xuena, Li Shiguo, Zhan Aibin. Environmental DNA (eDNA)-metabarcoding-based early monitoring and warning for invasive species in aquatic ecosystems [J]. Biodiv Sci, 2019, 27(5): 491-504. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn