



Biodiv Sci ›› 2023, Vol. 31 ›› Issue (7): 23073. DOI: 10.17520/biods.2023073 cstr: 32101.14.biods.2023073
• Original Papers: Animal Diversity • Previous Articles Next Articles
Zhiyuan Dong1,2, Linlin Chen1,2, Naipeng Zhang4, Li Chen1,3, Debin Sun1,2, Yanmei Ni1,3, Baoquan Li1,2,*( )
)
Received:2023-03-09
Accepted:2023-04-18
Online:2023-07-20
Published:2023-04-26
Contact:
*E-mail: Zhiyuan Dong, Linlin Chen, Naipeng Zhang, Li Chen, Debin Sun, Yanmei Ni, Baoquan Li. Response of fish diversity to hydrological connectivity of typical tidal creek system in the Yellow River Delta based on environmental DNA metabarcoding[J]. Biodiv Sci, 2023, 31(7): 23073.
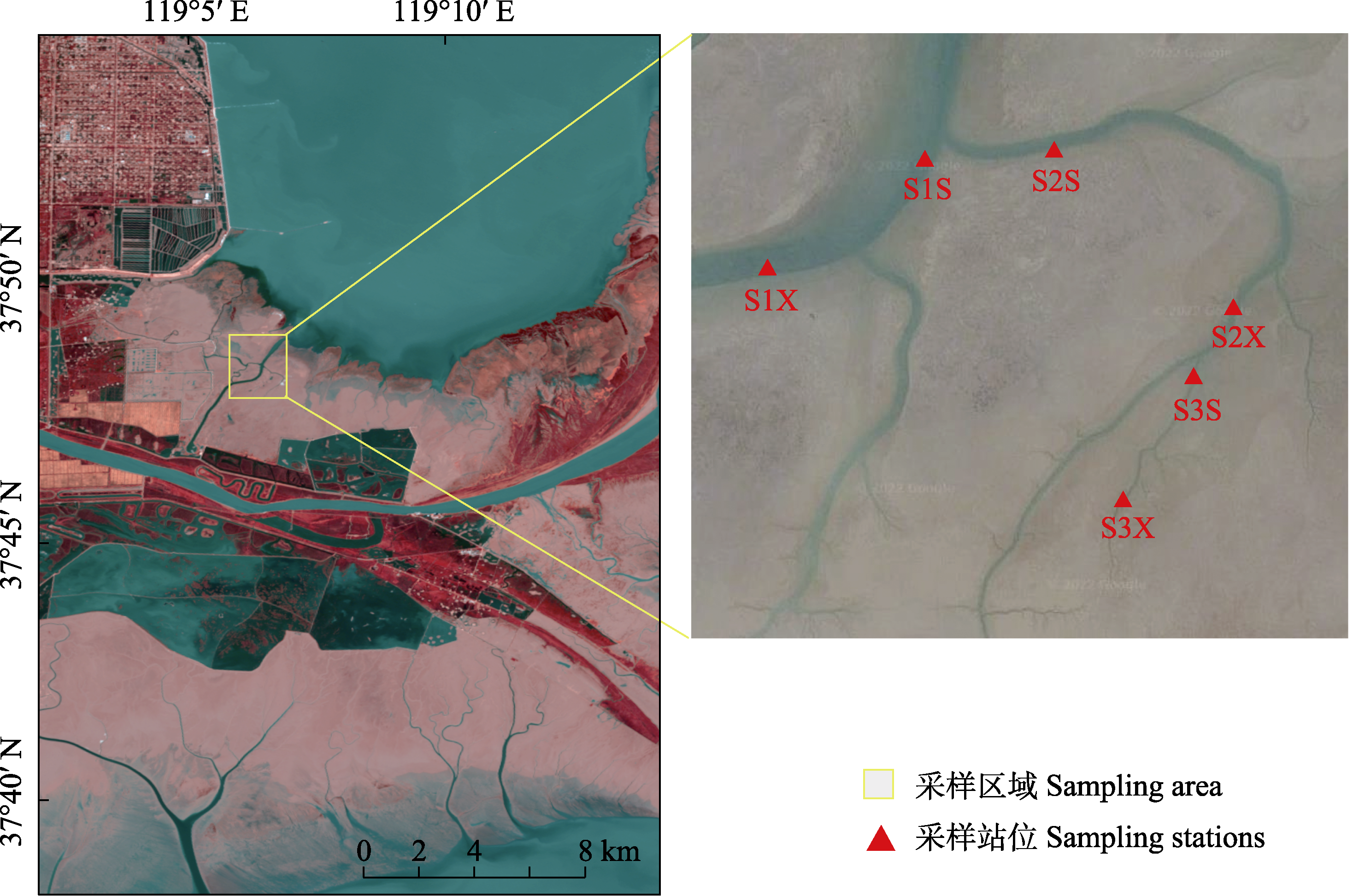
Fig. 1 Sampling area of natural wetland in the Yellow River Delta. S1S, Upstream of the first-class tidal creek; S1X, Downstream of the first-class tidal creek; S2S, Upstream of the second-class tidal creek; S2X, Downstream of the second-class tidal creek; S3S, Upstream of the third-class tidal creek; S3X, Downstream of the third-class tidal creek.
| 变量 Variable | 潮沟级别 Tidal creek level | ||
|---|---|---|---|
| 一级潮沟 The first-class tidal creek (S1) | 二级潮沟 The second-class tidal creek (S2) | 三级潮沟 The third-class tidal creek (S3) | |
| 总氮 TN (μg/L) | 351.893 ± 4.857b | 361.896 ± 11.934ab | 389.191 ± 19.168a |
| 总磷 TP (μg/L) | 15.116 ± 1.175a | 12.586 ± 0.580b | 14.975 ± 0.715a |
| 磷酸盐 PO43--P (mg/L) | 0.033 ± 0.029a | 0.008 ± 0.002ab | 0.004 ± 0.001b |
| 铵盐 NH4+-N (mg/L) | 0.039 ± 0.012a | 0.036 ± 0.004a | 0.170 ± 0.121b |
| 亚硝酸盐 NO2--N (mg/L) | 0.019 ± 0.003b | 0.024 ± 0.004ab | 0.027 ± 0.004a |
| 硅酸盐 SiO32--Si (mg/L) | 0.675 ± 0.052b | 0.690 ± 0.107ab | 0.964 ± 0.222a |
| 硝酸盐 NO3--N (mg/L) | 0.265 ± 0.041a | 0.238 ± 0.018a | 0.193 ± 0.017b |
| 温度 T (℃) | 21.800 ± 1.595b | 21.450 ± 0.321b | 26.233 ± 2.850a |
| 溶解氧 DO (mg/L) | 7.998 ± 0.804a | 6.692 ± 0.315b | 6.067 ± 1.143b |
| 电导率 SPC (ms/cm) | 26.412 ± 0.245b | 32.538 ± 1.752ab | 36.300 ± 1.715a |
| 盐度 SAL | 16.185 ± 0.170b | 20.362 ± 1.202ab | 22.903 ± 1.121a |
| 酸碱度 pH | 8.408 ± 0.012a | 8.385 ± 0.021ab | 8.248 ± 0.026b |
| 氧化还原电位 Eh | -71.950 ± 1.104a | -70.583 ± 1.087a | -64.033 ± 1.219b |
Table 1 Physical and chemical indicators of in the tidal creek of the Yellow River Estuary. Different lowercase letters in the same row meant significant difference at 0.05 level. Bolded numbers indicate maximum values. TN, Total nitrogen; TP: Total phosphorus; T: Temperature; DO: Dissolved oxygen; SPC: Conductivity; SAL: Salinity; Eh: Oxidation-reduction potential.
| 变量 Variable | 潮沟级别 Tidal creek level | ||
|---|---|---|---|
| 一级潮沟 The first-class tidal creek (S1) | 二级潮沟 The second-class tidal creek (S2) | 三级潮沟 The third-class tidal creek (S3) | |
| 总氮 TN (μg/L) | 351.893 ± 4.857b | 361.896 ± 11.934ab | 389.191 ± 19.168a |
| 总磷 TP (μg/L) | 15.116 ± 1.175a | 12.586 ± 0.580b | 14.975 ± 0.715a |
| 磷酸盐 PO43--P (mg/L) | 0.033 ± 0.029a | 0.008 ± 0.002ab | 0.004 ± 0.001b |
| 铵盐 NH4+-N (mg/L) | 0.039 ± 0.012a | 0.036 ± 0.004a | 0.170 ± 0.121b |
| 亚硝酸盐 NO2--N (mg/L) | 0.019 ± 0.003b | 0.024 ± 0.004ab | 0.027 ± 0.004a |
| 硅酸盐 SiO32--Si (mg/L) | 0.675 ± 0.052b | 0.690 ± 0.107ab | 0.964 ± 0.222a |
| 硝酸盐 NO3--N (mg/L) | 0.265 ± 0.041a | 0.238 ± 0.018a | 0.193 ± 0.017b |
| 温度 T (℃) | 21.800 ± 1.595b | 21.450 ± 0.321b | 26.233 ± 2.850a |
| 溶解氧 DO (mg/L) | 7.998 ± 0.804a | 6.692 ± 0.315b | 6.067 ± 1.143b |
| 电导率 SPC (ms/cm) | 26.412 ± 0.245b | 32.538 ± 1.752ab | 36.300 ± 1.715a |
| 盐度 SAL | 16.185 ± 0.170b | 20.362 ± 1.202ab | 22.903 ± 1.121a |
| 酸碱度 pH | 8.408 ± 0.012a | 8.385 ± 0.021ab | 8.248 ± 0.026b |
| 氧化还原电位 Eh | -71.950 ± 1.104a | -70.583 ± 1.087a | -64.033 ± 1.219b |
| 样品名称 Sample name | 序列条数 Sequences | 碱基数 Bases (bp) | 平均读长 Average length (bp) |
|---|---|---|---|
| S1S | 500,311 | 85,254,665 | 170.40 |
| S1X | 500,256 | 84,418,677 | 168.75 |
| S2S | 471,672 | 80,731,476 | 171.16 |
| S2X | 479,424 | 81,634,922 | 170.28 |
| S3S | 556,415 | 93,868,748 | 168.70 |
| S3X | 614,037 | 103,314,806 | 168.26 |
Table 2 Statistics of trimmed sequences in each sample
| 样品名称 Sample name | 序列条数 Sequences | 碱基数 Bases (bp) | 平均读长 Average length (bp) |
|---|---|---|---|
| S1S | 500,311 | 85,254,665 | 170.40 |
| S1X | 500,256 | 84,418,677 | 168.75 |
| S2S | 471,672 | 80,731,476 | 171.16 |
| S2X | 479,424 | 81,634,922 | 170.28 |
| S3S | 556,415 | 93,868,748 | 168.70 |
| S3X | 614,037 | 103,314,806 | 168.26 |
| 目 Order | 科 Family | 种 Species | |
|---|---|---|---|
| 本地鱼类物种 | 鲽形目 Pleuronectiformes | 舌鳎科 Cynoglossidae | 短吻红舌鳎 Cynoglossus joyneri |
| Native fish species | 鲱形目 Clupeiformes | 鲱科 Dorosomatidae | 青鳞小沙丁鱼 Sardinella zunasi |
| 斑鰶 Konosirus punctatus | |||
| 鳀科 Engraulidae | 赤鼻棱鳀 Thryssa kammalensis | ||
| 鳀 Engraulis japonicus | |||
| 凤鲚 Coilia mystus | |||
| 颌针鱼目 Beloniforme | 鱵科 Hemiramphidae | 间下鱵 Hyporhamphus intermedius | |
| 胡瓜鱼目 Osmeriformes | 胡瓜鱼科 Osmeridae | 西太公鱼 Hypomesus nipponensis | |
| 鲈形目 Perciformes | 花鲈科 Lateolabracidae | 花鲈 Lateolabrax japonicus | |
| 蓝子鱼科 Siganidae | 褐蓝子鱼 Siganus fuscescens | ||
| 鲹科 Carangidae | 吉打副叶鲹 Alepes djedaba | ||
| 石首鱼科 Sciaenidae | 小黄鱼 Larimichthys polyactis | ||
| 鱚科 Sillaginidae | 少鳞鱚 Sillago japonica | ||
| 虾虎鱼科 Gobiidae | 矛尾刺虾虎鱼 Acanthogobius hasta | ||
| 长体刺虾虎鱼 Acanthogobius elongatus | |||
| 矛尾虾虎鱼 Chaeturichthys stigmatias | |||
| 七棘裸身虾虎鱼 Gymnogobius heptacanthus | |||
| 普氏细棘虾虎鱼 Acentrogobius pflaumii | |||
| 六丝钝尾虾虎鱼 Amblychaeturichthys hexanema | |||
| 棕刺虾虎鱼 Acanthogobius luridus | |||
| 拉氏狼牙虾虎鱼 Odontamblyopus lacepedii | |||
| 鳗鲡目 Anguilliformes | 康吉鳗科 Congridae | 星康吉鳗 Conger myriaster | |
| 鳐形目 Rajiformes | 鳐科 Rajidae | 斑鳐 Okamejei kenojei | |
| 鲉形目 Scorpaeniformes | 六线鱼科 Hexagrammidae | 大泷六线鱼 Hexagrammos otakii | |
| 鲬科 Platycephalidae | 鲬 Platycephalus indicus | ||
| 鲻形目 Mugiliformes | 鲻科 Mugilidae | 鮻 Planiliza haematocheilus | |
| 鲻 Mugil cephalus | |||
| 非本地鱼类物种 | 鲱形目 Clupeiformes | 鲱科 Clupeidae | 隆背小沙丁鱼 Sardinella gibbosa |
| Non-native fish species | 花点鲥 Hilsa kelee | ||
| 鳉形目 Cyprinodontiformes | 青鳉科 Oryziatidae | 青鳉 Oryzias latipes | |
| 鲤形目 Cypriniformes | 鲤科 Cyprinidae | 鲫 Carassius auratus | |
| 似鳊 Pseudobrama simoni | |||
| 尖棘鱲 Zacco acutipinnis | |||
| 鲤 Cyprinus carpio | |||
| 温州光唇鱼 Acrossocheilus wenchowensis | |||
| 三角鲂 Megalobrama terminalis | |||
| 长鳍马口鱼 Opsariichthys evolans | |||
| 刺鲃 Spinibarbus caldwelli | |||
| 贝氏? Hemiculter bleekeri | |||
| 爬鳅科 Balitoridae | 梅花山拟腹吸鳅 Pseudogastromyzon meihuashanensis | ||
| 原缨口鳅 Vanmanenia stenosoma | |||
| 鲈形目 Perciformes | 金线鱼科 Nemipteridae | 日本金线鱼 Nemipterus thosaporni | |
| 丽鱼科 Cichlidae | 齐氏非鲫 Coptodon zillii | ||
| 非鲫 Oreochromis mossambicus | |||
| 加利略帚齿非鲫 Sarotherodon galilaeus | |||
| 尼罗非鲫 Oreochromis niloticus | |||
| 雀鲷科 Pomacentridae | 白带金翅雀鲷 Chrysiptera brownriggii | ||
| 塘鳢科 Eleotridae | 刺盖塘鳢 Eleotris acanthopomus | ||
| 虾虎鱼科 Gobiidae | 波氏吻虾虎鱼 Rhinogobius cliffordpopei | ||
| 雀斑吻虾虎鱼 Rhinogobius lentiginis | |||
| 名古屋吻虾虎鱼 Rhinogobius nagoyae | |||
| 武义吻虾虎鱼 Rhinogobius wuyiensis | |||
| 鲇形目 Siluriformes | 鲿科 Bagridae | 黄颡鱼 Tachysurus fulvidraco | |
| 鲇科 Siluridae | 怀头鲇 Silurus soldatovi | ||
| 甲鲇科 Loricariidae | 豹纹脂身鲇 Pterygoplichthys pardalis |
Table 3 List of fish species detected by eDNA metabarcoding in the tidal creek of the Yellow River Estuary. The term “Native fish species” refers to those marine fish that are actually caught or recorded as having been caught during the historical fishery resources survey in the Yellow River Estuary and Laizhou Bay.
| 目 Order | 科 Family | 种 Species | |
|---|---|---|---|
| 本地鱼类物种 | 鲽形目 Pleuronectiformes | 舌鳎科 Cynoglossidae | 短吻红舌鳎 Cynoglossus joyneri |
| Native fish species | 鲱形目 Clupeiformes | 鲱科 Dorosomatidae | 青鳞小沙丁鱼 Sardinella zunasi |
| 斑鰶 Konosirus punctatus | |||
| 鳀科 Engraulidae | 赤鼻棱鳀 Thryssa kammalensis | ||
| 鳀 Engraulis japonicus | |||
| 凤鲚 Coilia mystus | |||
| 颌针鱼目 Beloniforme | 鱵科 Hemiramphidae | 间下鱵 Hyporhamphus intermedius | |
| 胡瓜鱼目 Osmeriformes | 胡瓜鱼科 Osmeridae | 西太公鱼 Hypomesus nipponensis | |
| 鲈形目 Perciformes | 花鲈科 Lateolabracidae | 花鲈 Lateolabrax japonicus | |
| 蓝子鱼科 Siganidae | 褐蓝子鱼 Siganus fuscescens | ||
| 鲹科 Carangidae | 吉打副叶鲹 Alepes djedaba | ||
| 石首鱼科 Sciaenidae | 小黄鱼 Larimichthys polyactis | ||
| 鱚科 Sillaginidae | 少鳞鱚 Sillago japonica | ||
| 虾虎鱼科 Gobiidae | 矛尾刺虾虎鱼 Acanthogobius hasta | ||
| 长体刺虾虎鱼 Acanthogobius elongatus | |||
| 矛尾虾虎鱼 Chaeturichthys stigmatias | |||
| 七棘裸身虾虎鱼 Gymnogobius heptacanthus | |||
| 普氏细棘虾虎鱼 Acentrogobius pflaumii | |||
| 六丝钝尾虾虎鱼 Amblychaeturichthys hexanema | |||
| 棕刺虾虎鱼 Acanthogobius luridus | |||
| 拉氏狼牙虾虎鱼 Odontamblyopus lacepedii | |||
| 鳗鲡目 Anguilliformes | 康吉鳗科 Congridae | 星康吉鳗 Conger myriaster | |
| 鳐形目 Rajiformes | 鳐科 Rajidae | 斑鳐 Okamejei kenojei | |
| 鲉形目 Scorpaeniformes | 六线鱼科 Hexagrammidae | 大泷六线鱼 Hexagrammos otakii | |
| 鲬科 Platycephalidae | 鲬 Platycephalus indicus | ||
| 鲻形目 Mugiliformes | 鲻科 Mugilidae | 鮻 Planiliza haematocheilus | |
| 鲻 Mugil cephalus | |||
| 非本地鱼类物种 | 鲱形目 Clupeiformes | 鲱科 Clupeidae | 隆背小沙丁鱼 Sardinella gibbosa |
| Non-native fish species | 花点鲥 Hilsa kelee | ||
| 鳉形目 Cyprinodontiformes | 青鳉科 Oryziatidae | 青鳉 Oryzias latipes | |
| 鲤形目 Cypriniformes | 鲤科 Cyprinidae | 鲫 Carassius auratus | |
| 似鳊 Pseudobrama simoni | |||
| 尖棘鱲 Zacco acutipinnis | |||
| 鲤 Cyprinus carpio | |||
| 温州光唇鱼 Acrossocheilus wenchowensis | |||
| 三角鲂 Megalobrama terminalis | |||
| 长鳍马口鱼 Opsariichthys evolans | |||
| 刺鲃 Spinibarbus caldwelli | |||
| 贝氏? Hemiculter bleekeri | |||
| 爬鳅科 Balitoridae | 梅花山拟腹吸鳅 Pseudogastromyzon meihuashanensis | ||
| 原缨口鳅 Vanmanenia stenosoma | |||
| 鲈形目 Perciformes | 金线鱼科 Nemipteridae | 日本金线鱼 Nemipterus thosaporni | |
| 丽鱼科 Cichlidae | 齐氏非鲫 Coptodon zillii | ||
| 非鲫 Oreochromis mossambicus | |||
| 加利略帚齿非鲫 Sarotherodon galilaeus | |||
| 尼罗非鲫 Oreochromis niloticus | |||
| 雀鲷科 Pomacentridae | 白带金翅雀鲷 Chrysiptera brownriggii | ||
| 塘鳢科 Eleotridae | 刺盖塘鳢 Eleotris acanthopomus | ||
| 虾虎鱼科 Gobiidae | 波氏吻虾虎鱼 Rhinogobius cliffordpopei | ||
| 雀斑吻虾虎鱼 Rhinogobius lentiginis | |||
| 名古屋吻虾虎鱼 Rhinogobius nagoyae | |||
| 武义吻虾虎鱼 Rhinogobius wuyiensis | |||
| 鲇形目 Siluriformes | 鲿科 Bagridae | 黄颡鱼 Tachysurus fulvidraco | |
| 鲇科 Siluridae | 怀头鲇 Silurus soldatovi | ||
| 甲鲇科 Loricariidae | 豹纹脂身鲇 Pterygoplichthys pardalis |
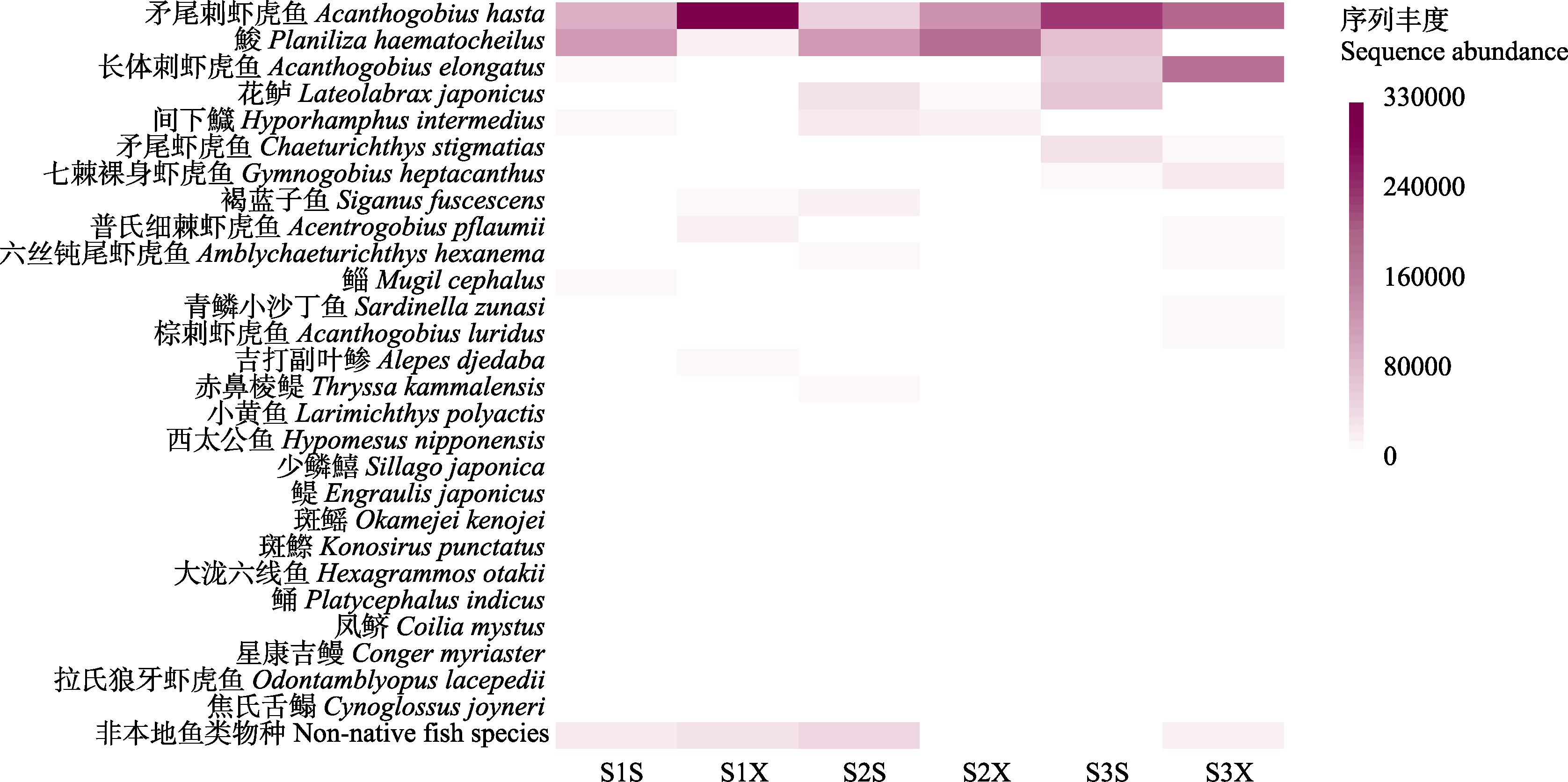
Fig. 3 Heatmap of the spatial distribution of fish species at each sampling station based on the abundance of sequences. The full names of the sampling stations' abbreviations are shown in Fig. 1.
| 采样站点 Sampling station | 观察物种数 Observed species | ACE指数 ACE index | Chao 1指数 Chao 1 index | Shannon指数 Shannon index | Simpson指数 Simpson index | 测序深度指数 Good's coverage |
|---|---|---|---|---|---|---|
| S1S | 325 | 328.28 | 328.11 | 3.95 | 0.84 | 0.999977 |
| S1X | 327 | 329.57 | 328.62 | 3.14 | 0.62 | 0.999980 |
| S2S | 324 | 335.00 | 350.25 | 4.44 | 0.87 | 0.999958 |
| S2X | 316 | 325.00 | 323.50 | 3.25 | 0.73 | 0.999958 |
| S3S | 325 | 337.51 | 335.69 | 3.60 | 0.78 | 0.999946 |
| S3X | 288 | 302.85 | 302.88 | 3.64 | 0.81 | 0.999935 |
Table 4 Alpha diversity index of fish community based on the relative abundance of sequences
| 采样站点 Sampling station | 观察物种数 Observed species | ACE指数 ACE index | Chao 1指数 Chao 1 index | Shannon指数 Shannon index | Simpson指数 Simpson index | 测序深度指数 Good's coverage |
|---|---|---|---|---|---|---|
| S1S | 325 | 328.28 | 328.11 | 3.95 | 0.84 | 0.999977 |
| S1X | 327 | 329.57 | 328.62 | 3.14 | 0.62 | 0.999980 |
| S2S | 324 | 335.00 | 350.25 | 4.44 | 0.87 | 0.999958 |
| S2X | 316 | 325.00 | 323.50 | 3.25 | 0.73 | 0.999958 |
| S3S | 325 | 337.51 | 335.69 | 3.60 | 0.78 | 0.999946 |
| S3X | 288 | 302.85 | 302.88 | 3.64 | 0.81 | 0.999935 |
| 排序轴 Axes | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 特征值 Eigenvalues | 0.62 | 0.251 | 0.046 | 0.028 |
| 相关系数 Species-environment correlations | 0.969 | 0.992 | 0.986 | 0.947 |
| 物种数据累积百分比方差 Cumulative percentage variance of species data (%) | 62 | 87.2 | 91.8 | 94.7 |
| 物种-环境关系累积百分比方差 Cumulative percentage variance of species-environment relation (%) | 65.3 | 91.7 | 96.6 | 99.6 |
| 总特征值 Sum of all eigenvalues | 1 | |||
| 总典范特征值 Sum of all canonical eigenvalues | 0.951 | |||
Table 5 Redundancy analysis (RDA) of dominant fish species in the tidal creek waters of Yellow River Estuary
| 排序轴 Axes | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 特征值 Eigenvalues | 0.62 | 0.251 | 0.046 | 0.028 |
| 相关系数 Species-environment correlations | 0.969 | 0.992 | 0.986 | 0.947 |
| 物种数据累积百分比方差 Cumulative percentage variance of species data (%) | 62 | 87.2 | 91.8 | 94.7 |
| 物种-环境关系累积百分比方差 Cumulative percentage variance of species-environment relation (%) | 65.3 | 91.7 | 96.6 | 99.6 |
| 总特征值 Sum of all eigenvalues | 1 | |||
| 总典范特征值 Sum of all canonical eigenvalues | 0.951 | |||
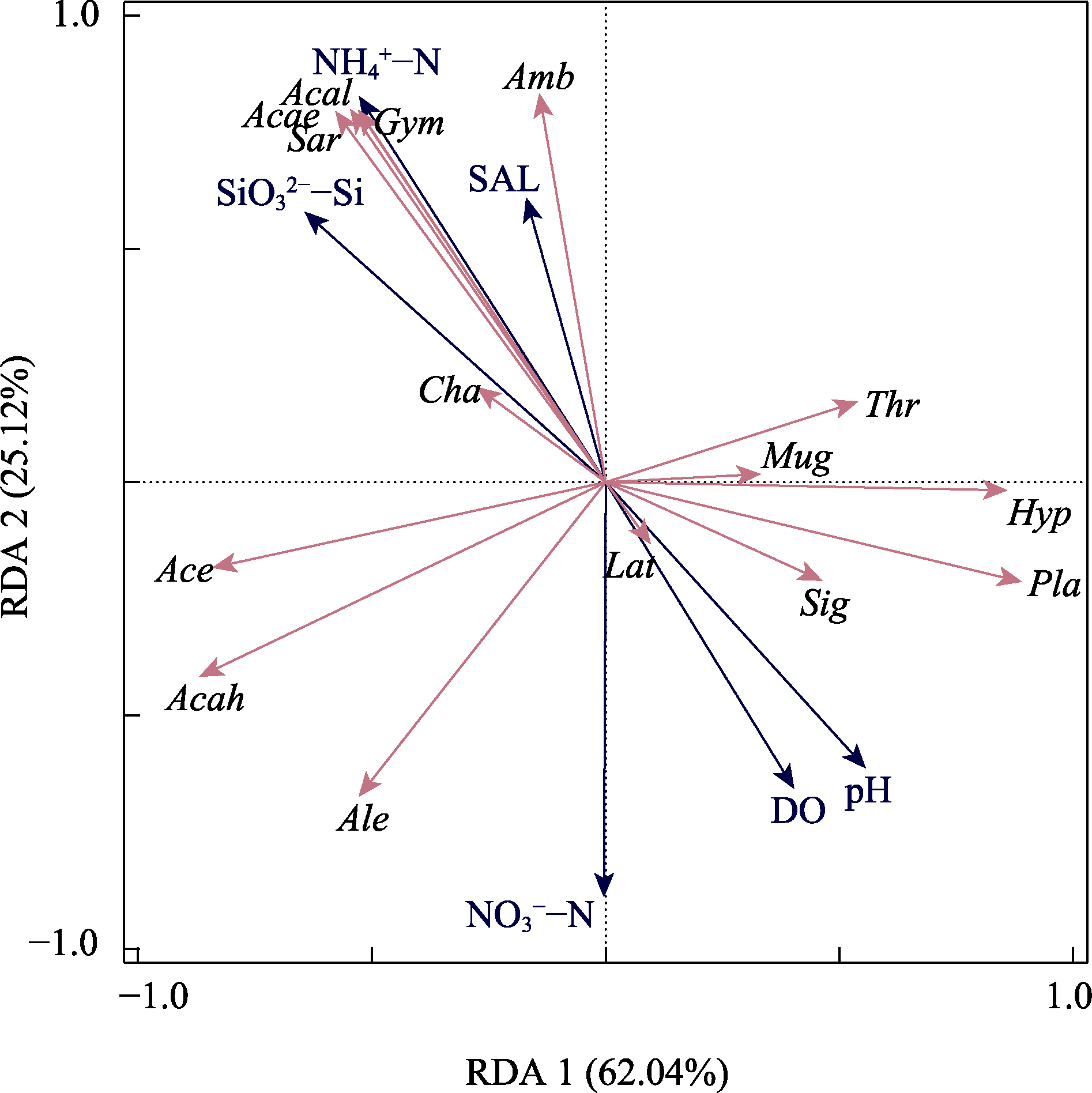
Fig. 4 Redundancy analysis (RDA) exploring the relationship between fish community structure and measured environmental variables. Acae, Acanthogobius elongatus; Acah, Acanthogobius hasta; Acal, Acanthogobius luridus; Ace, Acentrogobius pflaumii; Ale, Alepes djedaba; Amb, Amblychaeturichthys hexanema; Cha, Chaeturichthys stigmatias; Gym, Gymnogobius heptacanthus; Hyp, Hyporhamphus intermedius; Lat, Lateolabrax japonicus; Mug, Mugil cephalus; Pla, Planiliza haematocheilus; Sar, Sardinella zunasi; Sig, Siganus fuscescens; Thr, Thryssa kammalensis. DO, dissolved oxygen; NH4+-N, Ammoniumate; NO3--N, Nitrate; pH, Potential of hydrogen; SAL, Salinity; SiO32--Si, Silicate.
| [1] |
Balasingham KD, Walter RP, Mandrak NE, Heath DD (2018) Environmental DNA detection of rare and invasive fish species in two Great Lakes tributaries. Molecular Ecology, 27, 112-127.
DOI PMID |
| [2] | Chen WF, Zhou WZ, Shi YX (2003) Crisis of wetlands in the Yellow River Delta and its protection. Journal of Agro-Environmental Science, 22, 499-502. (in Chinese with English abstract) |
| [陈为峰, 周维芝, 史衍玺 (2003) 黄河三角洲湿地面临的问题及其保护. 农业环境科学学报, 22, 499-502.] | |
| [3] |
Connell JH (1978) High diversity of trees and corals is maintained only in a nonequilibrium state. Science, 199, 1302-1310.
DOI PMID |
| [4] |
Creer S, Deiner K, Frey S, Porazinska D, Taberlet P, Thomas WK, Potter C, Bik HM (2016) The ecologist's field guide to sequence-based identification of biodiversity. Methods in Ecology and Evolution, 7, 1008-1018.
DOI URL |
| [5] | Cui BS, Cai YZ, Xie T, Ning ZH, Hua YY (2016) Ecological effects of wetland hydrological connectivity: Problems and prospects. Journal of Beijing Normal University (Natural Science), 52, 738-746. (in Chinese with English abstract) |
| [崔保山, 蔡燕子, 谢湉, 宁中华, 华妍妍 (2016) 湿地水文连通的生态效应研究进展及发展趋势. 北京师范大学学报(自然科学版), 52, 738-746.] | |
| [6] |
Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière- Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872-5895.
DOI PMID |
| [7] | Dong R, Wang YY, Lü C, Lei GC, Xue BL, Chen QK (2020) Effects of hydrological connectivity on the community structure of macrobenthos in West Dongting Lake. Acta Ecologica Sinica, 40, 8336-8346. (in Chinese with English abstract) |
| [董芮, 王玉玉, 吕偲, 雷光春, 薛彬林, 陈乾阔 (2020) 水文连通性对西洞庭湖大型底栖动物群落结构的影响. 生态学报, 40, 8336-8346.] | |
| [8] |
Evangelisti M, D'Amelia D, Di Lallo GD, Thaller MC, Migliore L (2013) The relationship between salinity and bacterioplankton in three relic coastal ponds (Macchiatonda Wetland, Italy). Journal of Water Resource and Protection, 5, 859-866.
DOI URL |
| [9] |
Evans NT, Olds BP, Renshaw MA, Turner CR, Li YY, Jerde CL, Mahon AR, Pfrender ME, Lamberti GA, Lodge DM (2016) Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Molecular Ecology Resources, 16, 29-41.
DOI PMID |
| [10] | Fan HY, Ji YP, Zhang SH, Yuan CT, Gao TX (2005) Research of fishery biology of the neritic fish Synechogobius ommaturus in the area of the Huanghe Delta. Periodical of Ocean University of China (Natural Science), 35, 733-736. (in Chinese with English abstract) |
| [范海洋, 纪毓鹏, 张士华, 苑春亭, 高天翔 (2005) 黄河三角洲斑尾复虾虎鱼渔业生物学的研究. 中国海洋大学学报(自然科学版), 35, 733-736.] | |
| [11] |
Freeman MC, Pringle CM, Jackson CR (2007) Hydrologic connectivity and the contribution of stream headwaters to ecological integrity at regional scales. Journal of the American Water Resources Association, 43, 5-14.
DOI URL |
| [12] | Gao XC, Jiang W (2021) The construction and application of BLAST database of DNA barcode for common fish in the Three Gorges Reservoir. Genomics and Applied Biology, 40, 1952-1960. (in Chinese with English abstract) |
| [郜星辰, 姜伟 (2021) 三峡库区常见鱼类DNA条形码本地BLAST数据库的构建和应用. 基因组学与应用生物学, 40, 1952-1960.] | |
| [13] |
Garwood JA, Allen DM, Kimball ME, Boswell KM (2019) Site fidelity and habitat use by young-of-the-year transient fishes in salt marsh intertidal creeks. Estuaries and Coasts, 42, 1387-1396.
DOI |
| [14] |
Jardine TD, Pettit NE, Warfe DM, Pusey BJ, Ward DP, Douglas MM, Davies PM, Bunn SE (2012) Consumer-resource coupling in wet-dry tropical rivers. Journal of Animal Ecology, 81, 310-322.
DOI PMID |
| [15] |
Ji FF, Han DY, Yan L, Yan SH, Zha JM, Shen JZ (2022) Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method. Science of the Total Environment, 806, 150587.
DOI URL |
| [16] | Jiang PW, Li M, Zhang S, Chen ZZ, Xu SN (2022) Investigating the fish diversity in Pearl River Estuary based on environmental DNA metabarcoding and bottom trawling. Acta Hydrobiologica Sinica, 46, 1701-1711. (in Chinese with English abstract) |
| [蒋佩文, 李敏, 张帅, 陈作志, 徐姗楠 (2022) 基于环境DNA宏条码和底拖网的珠江河口鱼类多样性. 水生生物学报, 46, 1701-1711.] | |
| [17] | Jiang WZ, Sui KG (2015) Study on ecological pond aquaculture technology of Tachysurus fulvidraco in saline-alkali land. Hebei Fisheries, (11), 32-33. (in Chinese) |
| [蒋万钊, 隋凯港 (2015) 盐碱地黄颡鱼池塘生态养殖技术研究. 河北渔业, (11), 32-33.] | |
| [18] |
Jo TS (2023) Correlation between the number of eDNA particles and species abundance is strengthened by warm temperature: Simulation and meta-analysis. Hydrobiologia, 850, 39-50.
DOI |
| [19] |
Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZJ, Zaneveld JR, Zhu QY, Caporaso JG, Dorrestein PC (2018) Best practices for analysing microbiomes. Nature Reviews Microbiology, 16, 410-422.
DOI PMID |
| [20] |
Kume M, Lavergne E, Ahn H, Terashima Y, Kadowaki K, Ye F, Kameyama S, Kai Y, Henmi YM, Yamashita Y, Kasai A (2021) Factors structuring estuarine and coastal fish communities across Japan using environmental DNA metabarcoding. Ecological Indicators, 121, 107216.
DOI URL |
| [21] |
Lamb PD, Fonseca VG, Maxwell DL, Nnanatu CC (2022) Systematic review and meta-analysis: Water type and temperature affect environmental DNA decay. Molecular Ecology Resources, 22, 2494-2505.
DOI PMID |
| [22] |
Li DX, Li YQ, Zhang KH, Ma X, Zhang SY, Liu WH, Che CG, Cui BS (2020) Characteristics of carbon and nitrogen distribution in typical tidal creeks of the Yellow River Delta. Journal of Natural Resources, 35, 460-471. (in Chinese with English abstract)
DOI URL |
|
[李冬雪, 李雨芩, 张珂豪, 马旭, 张树岩, 刘伟华, 车纯广, 崔保山 (2020) 黄河口典型潮沟土壤碳氮分布特征规律. 自然资源学报, 35, 460-471.]
DOI |
|
| [23] |
Li F, Lü ZB, Wei ZH, Wang TT, Xu BQ, Wang ZQ (2013) Seasonal changes in the community structure of the demersal fishery in Laizhou Bay. Journal of Fishery Sciences of China, 20, 137-147. (in Chinese with English abstract)
DOI URL |
| [李凡, 吕振波, 魏振华, 王田田, 徐炳庆, 王忠全 (2013) 2010年莱州湾底层渔业生物群落结构及季节变化. 中国水产科学, 20, 137-147.] | |
| [24] | Li F, Xu BQ, Lü ZB, Wang TT (2018) Ecological niche of dominant species of fish assemblages in Laizhou Bay, China. Acta Ecologica Sinica, 38, 5195-5205. (in Chinese with English abstract) |
| [李凡, 徐炳庆, 吕振波, 王田田 (2018) 莱州湾鱼类群落优势种生态位. 生态学报, 38, 5195-5205.] | |
| [25] | Li HT, Zou KS, Zhang S, Cao YT, Lu ZC, Chen ZZ, Li M (2022) Species composition of fishes in the Pearl River Estuary based on environmental DNA metabarcoding. Journal of Shanghai Ocean University, 31, 1423-1433. (in Chinese with English abstract) |
| [李红婷, 邹柯姝, 张帅, 曹漪婷, 卢芷程, 陈作志, 李敏 (2022) 基于环境DNA宏条形码的珠江河口鱼类种类组成. 上海海洋大学学报, 31, 1423-1433.] | |
| [26] | Li MD, Wang XL, Lü X (1997) Mullet. China Ocean Press, Beijing. (in Chinese) |
| [李明德, 王秀玲, 吕宪 (1997) 梭鱼. 海洋出版社, 北京.] | |
| [27] | Li W, Cui LJ, Zhao XS, Zhang MY, Ma MY, Kang XM, Wang YF (2014) An overview of Chinese coastal wetland and their ecosystem services. Forest Inventory and Planning, 39(4), 24-30. (in Chinese with English abstract) |
| [李伟, 崔丽娟, 赵欣胜, 张曼胤, 马牧源, 康晓明, 王义飞 (2014) 中国滨海湿地及其生态系统服务功能研究概述. 林业调查规划, 39(4), 24-30.] | |
| [28] | Liu J, Chen YX, Ma L (2015) Fishes of the Bohai Sea and Yellow Sea. Science Press, Beijing. (in Chinese) |
| [刘静, 陈咏霞, 马琳 (2015) 黄渤海鱼类图志. 科学出版社, 北京.] | |
| [29] | Luo M, Wang Q, Qiu DD, Shi W, Ning ZH, Cai YZ, Song ZF, Cui BS (2018) Hydrological connectivity characteristics and ecological effects of a typical tidal channel system in the Yellow River Delta. Journal of Beijing Normal University (Natural Science), 54, 17-24. (in Chinese with English abstract) |
| [骆梦, 王青, 邱冬冬, 施伟, 宁中华, 蔡燕子, 宋振峰, 崔保山 (2018) 黄河三角洲典型潮沟系统水文连通特征及其生态效应. 北京师范大学学报(自然科学版), 54, 17-24.] | |
| [30] | Ma QZ, Zhang TT, Zhao F, Zhang T, Yang G, Wang SK (2023) Effects of tidal creek connectivity on fish communities in the Yangtze estuary wetlands. Chinese Journal of Ecology, http://kns.cnki.net/kcms/detail/21.1148.Q.20230311.1446.006.html. (in Chinese with English abstract) |
| [马巧珍, 张婷婷, 赵峰, 张涛, 杨刚, 王思凯 (2023) 长江口湿地潮沟连通程度对鱼类群落的影响. 生态学杂志, http://kns.cnki.net/kcms/detail/21.1148.Q.20230311.1446.006.html.] | |
| [31] |
Miya M, Gotoh RO, Sado T (2020) MiFish metabarcoding: A high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples. Fisheries Science, 86, 939-970.
DOI |
| [32] |
Miya M, Nishida M (2000) Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximum-parsimony optimality criterion. Molecular Phylogenetics and Evolution, 17, 437-455.
PMID |
| [33] | Miya M, Sato Y, Fukunaga T, Sado T, Poulsen JY, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, Kondoh M, Iwasaki W (2015) MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. Royal Society Open Science, 2, 150088. |
| [34] |
Murakami H, Yoon S, Kasai A, Minamoto T, Yamamoto S, Sakata MK, Horiuchi T, Sawada H, Kondoh M, Yamashita Y, Masuda R (2019) Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fisheries Science, 85, 327-337.
DOI |
| [35] |
Nakagawa H, Yamamoto S, Sato Y, Sado T, Minamoto T, Miya M (2018) Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshwater Biology, 63, 569-580.
DOI URL |
| [36] | O'Donnell JL, Kelly RP, Shelton AO, Samhouri JF, Lowell NC, Williams GD (2017) Spatial distribution of environmental DNA in a nearshore marine habitat. PeerJ, 5, e3044. |
| [37] | Qu JY, Yang GMM, Fang Z, Chen XJ (2021) A review of research advancement on fisheries biology of Japanese Spanish Mackerel Scomberomorus niphonius. Fisheries Science, 40, 643-650. (in Chinese with English abstract) |
| [瞿俊跃, 杨光明媚, 方舟, 陈新军 (2021) 蓝点马鲛渔业生物学研究进展. 水产科学, 40, 643-650.] | |
| [38] |
Sales NG, Wangensteen OS, Carvalho DC, Deiner K, Præbel K, Coscia I, McDevitt AD, Mariani S (2021) Space-time dynamics in monitoring neotropical fish communities using eDNA metabarcoding. Science of the Total Environment, 754, 142096.
DOI URL |
| [39] |
Sard NM, Herbst SJ, Nathan L, Uhrig G, Kanefsky J, Robinson JD, Scribner KT (2019) Comparison of fish detections, community diversity, and relative abundance using environmental DNA metabarcoding and traditional gears. Environmental DNA, 1, 368-384.
DOI URL |
| [40] |
Schofield KA, Alexander LC, Ridley CE, Vanderhoof MK, Fritz KM, Autrey BC, DeMeester JE, Kepner WG, Lane CR, Leibowitz SG, Pollard AI (2018) Biota connect aquatic habitats throughout freshwater ecosystem mosaics. Journal of the American Water Resources Association, 54, 372-399.
DOI PMID |
| [41] |
Shaw JLA, Clarke LJ, Wedderburn SD, Barnes TC, Weyrich LS, Cooper A (2016) Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biological Conservation, 197, 131-138.
DOI URL |
| [42] |
Shen M, Guo NN, Luo ZL, Guo XC, Sun G, Xiao NW (2022) Explore the distribution and influencing factors of fish in major rivers in Beijing with eDNA metabarcoding technology. Biodiversity Science, 30, 22240. (in Chinese with English abstract)
DOI |
|
[沈梅, 郭宁宁, 罗遵兰, 郭晓晨, 孙光, 肖能文 (2022) 基于eDNA metabarcoding探究北京市主要河流鱼类分布及影响因素. 生物多样性, 30, 22240.]
DOI |
|
| [43] |
Sousa WP (1979) Disturbance in marine intertidal boulder fields: The nonequilibrium maintenance of species diversity. Ecology, 60, 1225-1239.
DOI URL |
| [44] |
Strickler KM, Fremier AK, Goldberg CS (2015) Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation, 183, 85-92.
DOI URL |
| [45] | Sun JF, Liu QZ (2010) Research on comprehensive development and utilization of geothermal resources in Dongying. Journal of Hebei Agricultural Sciences, 14(2), 91-93, 96. (in Chinese with English abstract) |
| [孙金凤, 刘清志 (2010) 东营市地热资源综合开发与利用研究. 河北农业科学, 14(2), 91-93, 96.] | |
| [46] | Sun PF, Shan XJ, Wu Q, Chen YL, Jin XS (2014) Seasonal variations in fish community structure in the Laizhou Bay and the Yellow River Estuary. Acta Ecologica Sinica, 34, 367-376. (in Chinese with English abstract) |
| [孙鹏飞, 单秀娟, 吴强, 陈云龙, 金显仕 (2014) 莱州湾及黄河口水域鱼类群落结构的季节变化. 生态学报, 34, 367-376.] | |
| [47] | Sun ZG, Mou XJ, Chen XB, Wang LL, Song HL, Jiang HH (2011) Actualities, problems and suggestions of wetland protection and restoration in the Yellow River Delta. Wetland Science, 9, 107-115. (in Chinese with English abstract) |
| [孙志高, 牟晓杰, 陈小兵, 王玲玲, 宋红丽, 姜欢欢 (2011) 黄河三角洲湿地保护与恢复的现状、问题与建议. 湿地科学, 9, 107-115.] | |
| [48] | Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E (2012a) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE, 7, e41732. |
| [49] |
Thomsen PF, Kielgast J, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP, Orlando L, Willerslev E (2012b) Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology, 21, 2565-2573.
DOI URL |
| [50] |
Tsuji S, Takahara T, Doi H, Shibata N, Yamanaka H,(2019) The detection of aquatic macroorganisms using environmental DNA analysis—A review of methods for collection, extraction, and detection. Environmental DNA, 1, 99-108.
DOI URL |
| [51] |
Valentini A, Taberlet P, Miaud C, Civade R, Herder J, Thomsen PF, Bellemain E, Besnard A, Coissac E, Boyer F, Gaboriaud C, Jean P, Poulet N, Roset N, Copp GH, Geniez P, Pont D, Argillier C, Baudoin JM, Peroux T, Crivelli AJ, Olivier A, Acqueberge M, Le Brun M, Møller PR, Willerslev E, Dejean T (2016) Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Molecular Ecology, 25, 929-942.
DOI PMID |
| [52] | Wang P, Jiao Y, Ren YP, Zhong CJ, Yu H (1999) Investigation of biodiversity from spring catch in coastal waters of Laizhou Bay and Huanghe Estuary. Transaction of Oceanology and Limnology, (1), 40-44. (in Chinese with English abstract) |
| [王平, 焦燕, 任一平, 仲崇俊, 于浩 (1999) 莱州湾、黄河口水域春季近岸渔获生物多样性特征的调查研究. 海洋湖沼通报, (1), 40-44.] | |
| [53] | Wang RX, Yang G, Geng Z, Zhao F, Feng XS, Zhang T (2023) Application of environmental DNA technology in fish diversity analysis in the Yangtze River estuary. Acta Hydrobiologica Sinica, 47, 365-375. (in Chinese with English abstract) |
| [王汝贤, 杨刚, 耿智, 赵峰, 冯雪松张涛 (2023) 环境DNA技术在长江口鱼类多样性分析中的应用. 水生生物学报, 47, 365-375.] | |
| [54] |
Wang XY, Yan JG, Bai JH, Cui BS (2019) Influence of hydrological connectivity of coastal wetland on the biological connectivity of macrobenthos in the Yellow River Estuary. Journal of Natural Resources, 34, 2544-2553. (in Chinese with English abstract)
DOI URL |
|
[王新艳, 闫家国, 白军红, 崔保山 (2019) 黄河口滨海湿地水文连通对大型底栖动物生物连通的影响. 自然资源学报, 34, 2544-2553.]
DOI |
|
| [55] |
Yamamoto S, Masuda R, Sato Y, Sado T, Araki H, Kondoh M, Minamoto T, Miya M (2017) Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Scientific Reports, 7, 40368.
DOI PMID |
| [56] | Yamamoto S, Minami K, Fukaya K, Takahashi K, Sawada H, Murakami H, Tsuji S, Hashizume H, Kubonaga S, Horiuchi T, Hongo M, Nishida J, Okugawa Y, Fujiwara A, Fukuda M, Hidaka S, Suzuki KW, Miya M, Araki H, Yamanaka H, Maruyama A, Miyashita K, Masuda R, Minamoto T, Kondoh M (2016) Environmental DNA as a ‘snapshot' of fish distribution: A case study of Japanese Jack Mackerel in Maizuru Bay, Sea of Japan. PLoS ONE, 11, e0149786. |
| [57] |
Yin S, Bai JH, Wang X, Wang XY, Zhang GL, Jia J, Li XW, Liu XH (2020) Hydrological connectivity and herbivores control the autochthonous producers of coastal salt marshes. Marine Pollution Bulletin, 160, 111638.
DOI URL |
| [58] |
Yuan J, Cohen MJ, Kaplan DA, Acharya S, Larsen LG, Nungesser MK (2015) Linking metrics of landscape pattern to hydrological process in a lotic wetland. Landscape Ecology, 30, 1893-1912.
DOI URL |
| [59] |
Zhan AB, Hulák M, Sylvester F, Huang XT, Adebayo AA, Abbott CL, Adamowicz SJ, Heath DD, Cristescu ME, MacIsaac HJ (2013) High sensitivity of 454 pyrosequencing for detection of rare species in aquatic communities. Methods in Ecology and Evolution, 4, 558-565.
DOI URL |
| [60] | Zhang DY, Chai XJ, Ruan ZC, Zhang Y, Wang YB (2020) Effects of long time low salinity on the growth and physiology in the juvenile of Liza haematocheila. Journal of Zhejiang Ocean University (Natural Science), 39, 296-302. (in Chinese with English abstract) |
| [张鼎元, 柴学军, 阮泽超, 张燕, 王跃斌 (2020) 长期低盐胁迫对梭鱼生长及生理的影响. 浙江海洋大学学报(自然科学版), 39, 296-302.] | |
| [61] | Zhang XX, Yao QZ, Chen HT, Mi TZ, Tan JQ, Yu ZG (2010) Seasonal variation and fluxes of nutrients in the lower reaches of the Yellow River. Periodical of Ocean University of China, 40(7), 82-88. (in Chinese with English abstract) |
| [张晓晓, 姚庆祯, 陈洪涛, 米铁柱, 谭加强, 于志刚 (2010) 黄河下游营养盐浓度季节变化及其入海通量研究. 中国海洋大学学报(自然科学版), 40(7), 82-88.] | |
| [62] | Zhao XY, Li X, Yu M, Yu JB, Wang XH (2019) Actualities of wetland hydrological connectivity blocked in the Yellow River Delta. Regional Governance, (49), 78-81. (in Chinese) |
| [赵心怡, 李晓, 于淼, 于君宝, 王雪宏 (2019) 黄河三角洲湿地水文连通受阻现状. 区域治理, (49), 78-81.] | |
| [63] |
Zou KS, Chen JW, Ruan HT, Li ZH, Guo WJ, Li M, Liu L (2020) eDNA metabarcoding as a promising conservation tool for monitoring fish diversity in a coastal wetland of the Pearl River Estuary compared to bottom trawling. Science of the Total Environment, 702, 134704.
DOI URL |
| [1] | Ma Wenjun, Liu Sijia, Li Kemao, Jian Shenglong, Xue Chang’an, Han Qingxiango, Wei Jinliang, Chen Shengxue, Niu Yimeng, Cui Zhouping, Sui Ruichen, Tian Fei, Zhao Kai. Fish diversity and distribution in the source region of the Yangtze River in Qinghai Province [J]. Biodiv Sci, 2025, 33(2): 24494-. |
| [2] | Zuopeng Zhang, Chenyang Yao, Ling Wu, Zunlan Luo, Guang Sun, Zongyong Guo, Xiaosi Li, Feng Lin, Xiaoyong Chen. Fish diversity and threat factors in the Yunnan section of the Nujiang River [J]. Biodiv Sci, 2024, 32(7): 24076-. |
| [3] | Yanmei Ni, Li Chen, Zhiyuan Dong, Debin Sun, Baoquan Li, Xumin Wang, Linlin Chen. Community structure of macrobenthos and ecological health evaluation in the restoration area of the Yellow River Delta wetland [J]. Biodiv Sci, 2024, 32(3): 23303-. |
| [4] | Xin Gao, Yahui Zhao, Fei Tian, Xiaoai Wang, Mingzheng Li, Pengcheng Lin, Tao Chang, Dan Yu, Huanzhang Liu. Progress of monitoring and research of China Inland Water Fish Biodiversity Observation Network [J]. Biodiv Sci, 2023, 31(12): 23427-. |
| [5] | Wei Zhou, Minghui Li, Youlan Li. Fish diversity in four nature reserves in Southwest Yunnan, China and the evaluation indicators [J]. Biodiv Sci, 2016, 24(3): 313-320. |
| [6] | Yang Junxing, Chen Yinrui, He Yuanhui. Studies on fish diversity in plateau lakes Of the central Yunnan [J]. Biodiv Sci, 1994, 02(4): 204-209. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()