



生物多样性 ›› 2019, Vol. 27 ›› Issue (5): 480-490. DOI: 10.17520/biods.2018227 cstr: 32101.14.biods.2018227
所属专题: 生物入侵
李萌1,尉婷婷1,史博洋1,郝希阳1,徐海根2,孙红英1,*( )
)
收稿日期:2018-08-17
接受日期:2018-12-27
出版日期:2019-05-20
发布日期:2019-05-20
通讯作者:
孙红英
基金资助:
Li Meng1,Wei Tingting1,Shi Boyang1,Hao Xiyang1,Xu Haigen2,Sun Hongying1,*( )
)
Received:2018-08-17
Accepted:2018-12-27
Online:2019-05-20
Published:2019-05-20
Contact:
Sun Hongying
摘要:
环境DNA (eDNA)是指生物有机体在环境中(例如土壤、沉积物或水体)遗留下的DNA片段。eDNA技术是指从环境中提取DNA片段进行测序以及数据分析来反映环境中的物种或群落信息。与传统方法相比, eDNA技术具有高灵敏度、省时省力、无损伤等优点。目前, eDNA技术已成为一种新的水生生物监测方法, 主要应用于水生生物的多样性研究、濒危和稀有动物的物种状态及外来入侵动物扩散动态的监测等。本文从eDNA技术在水生生物多样性监测应用领域的发展历程、eDNA技术的操作流程以及其在监测淡水底栖大型无脊椎动物方面的应用进展、技术优势和局限性五个方面进行了综述。最后, 本文对eDNA技术在淡水底栖大型无脊椎动物多样性监测应用的发展趋势和前景作出展望。
李萌, 尉婷婷, 史博洋, 郝希阳, 徐海根, 孙红英 (2019) 环境DNA技术在淡水底栖大型无脊椎动物多样性监测中的应用. 生物多样性, 27, 480-490.
DOI: 10.17520/biods.2018227.
Li Meng, Wei Tingting, Shi Boyang, Hao Xiyang, Xu Haigen, Sun Hongying (2019) Biodiversity monitoring of freshwater benthic macroinvertebrates using environmental DNA. Biodiversity Science, 27, 480-490. DOI: 10.17520/biods.2018227.
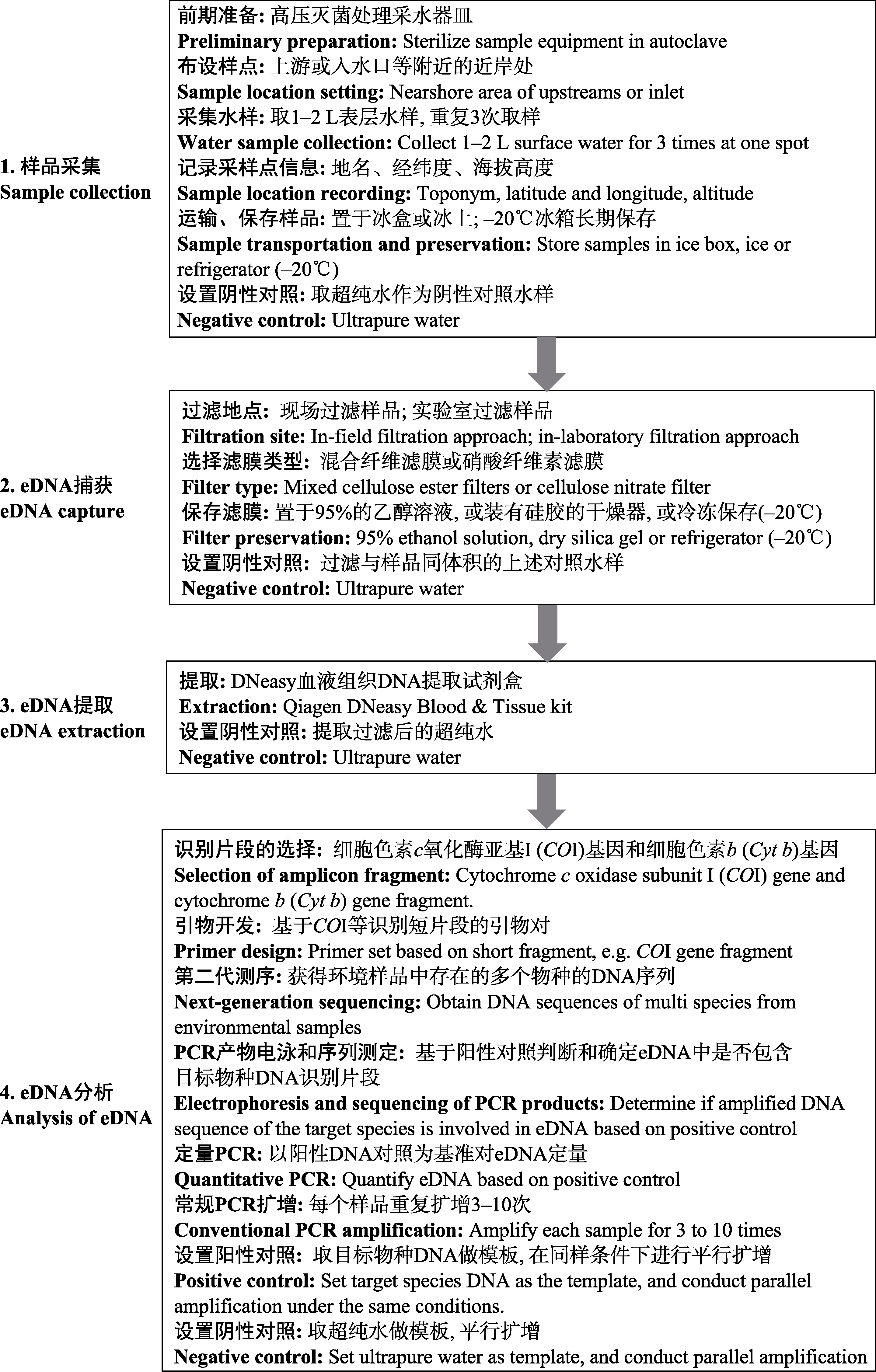
图1 利用eDNA宏条形码技术开展淡水底栖大型无脊椎动物多样性监测的主要流程
Fig. 1 The framework of biodiversity monitoring for freshwater benthic macroinvertebrates using eDNA metabarcoding
| [1] |
Andújar C, Arribas P, Yu DW, Vogler AP, Emerson BC ( 2018) Why the COI barcode should be the community DNA metabarcode for the metazoa. Molecular Ecology, 27, 3968-3975.
DOI URL |
| [2] | Balasingham KD, Walter RP, Heath DD ( 2016) Residual eDNA detection sensitivity assessed by quantitative real time PCR in a river ecosystem. Molecular Ecology Resources, 17, 523-532. |
| [3] | Cai FJ, Wu ZJ, He N, Ning L, Huang CM ( 2010) Research progress in invasion ecology of Procambarus clarkia. Chinese Journal of Ecology, 29, 124-132. (in Chinese with English abstract) |
| [ 蔡凤金, 武正军, 何南, 宁蕾, 黄乘明 ( 2010) 克氏原螯虾的入侵生态学研究进展. 生态学杂志, 29, 124-132.] | |
| [4] | Cai W, Ma Z, Yang C, Wang L, Wang W, Zhao G, Geng Y, Yu DW ( 2017) Using eDNA to detect the distribution and density of invasive crayfish in the Honghe-Hani Rice Terrace World Heritage Site. PLoS ONE, 12, e0177724. |
| [5] | Cao L, Zhang E, Zang CX, Cao WX ( 2016) Evaluating the status of China’s continental fish and analyzing their causes of endangerment through the red list assessment. Biodiversity Science, 24, 598-609. (in Chinese with English abstract) |
| [ 曹亮, 张鹗, 臧春鑫, 曹文宣 ( 2016) 通过红色名录评估研究中国内陆鱼类受威胁现状及其成因. 生物多样性, 24, 598-609.] | |
| [6] | Champlot S, Berthelot C, Pruvost M, Bennett EA, Grange T, Geigl EM ( 2010) An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE, 5, e13042. |
| [7] | Chu KL, Ma XP, Zhang ZW, Wang PF, Lv LN, Zhao Q, Sun HY ( 2018) A checklist for the classification and distribution of China’s freshwater crabs. Biodiversity Science, 26, 274-282. (in Chinese with English abstract) |
| [ 楚克林, 马晓萍, 张泽伟, 王鹏飞, 吕琳娜, 赵强, 孙红英 ( 2018) 中国淡水蟹分类与分布名录(十足目: 拟地蟹科, 溪蟹科). 生物多样性, 26, 274-282.] | |
| [8] | Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière- Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, Vere ND ( 2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872-5895. |
| [9] | Deiner K, Fronhofer EA, Mächler E, Walser JC, Altermatt F ( 2016) Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nature Communications, 7, 12544. |
| [10] | Deiner K, Walser JC, Mächler E, Altermatt F ( 2015) Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biological Conservation, 183, 53-63. |
| [11] | Deiner K, Altermatt F ( 2014) Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE, 9, e88786. |
| [12] | Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E, Miaud C ( 2012) Improved detection of an alien invasive species through environmental DNA barcoding: The example of the American bullfrog Lithobates catesbeianus. Journal of Applied Ecology, 49, 953-959. |
| [13] | Ding H, Ma FZ, Wu J, Lei JC, Le ZF, Xu HG ( 2015) Considerations of building up a supervision and management system for prevention and control of invasive alien species posing environmental hazards in China. Journal of Ecology and Rural Environment, 31, 652-657. (in Chinese with English abstract) |
| [ 丁晖, 马方舟, 吴军, 雷军成, 乐志芳, 徐海根 ( 2015) 关于构建我国外来入侵物种环境危害防控监督管理体系的思考. 生态与农村环境学报, 31, 652-657.] | |
| [14] | Doi H, Katano I, Sakata Y, Souma R, Kosuge T, Nagano M, Ikeda K, Yano K, Tojo K ( 2017) Detection of an endangered aquatic heteropteran using environmental DNA in a wetland ecosystem. Royal Society Open Science, 4, 170568. |
| [15] | Dougherty MM, Larson ER, Renshaw MA, Gantz CA, Egan SP, Erickson DM, Lodge DM ( 2016) Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. Journal of Applied Ecology, 53, 722-732. |
| [16] | Duan XH, Wang ZY, Xu MZ ( 2010) Benthic Macroinvertebrate and Application in the Assessment of Stream Ecology. Tsinghua University Press,Beijing. (in Chinese) |
| [ 段学花, 王兆印, 徐梦珍 ( 2010) 底栖动物与河流生态评价.清华大学出版社, 北京.] | |
| [17] | Egan SP, Grey E, Olds B, Feder JL, Ruggiero ST, Tanner CE, Lodge DM ( 2015) Rapid molecular detection of invasive species in ballast and harbor water by integrating environmental DNA and light transmission spectroscopy. Environmental Science & Technology, 49, 4113-4121. |
| [18] | Eichmiller JJ, Bajer PG, Sorensen PW ( 2014) The relationship between the distribution of common carp and their environmental DNA in a small lake. PLoS ONE, 9, e112611. |
| [19] | Eichmiller JJ, Miller LM, Sorensen PW ( 2016) Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish. Molecular Ecology Resources, 16, 56-68. |
| [20] | Elbrecht V, Leese F ( 2017) Validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment. Frontiers in Environmental Science, 5, 11. |
| [21] |
Fernández S, Rodríguez S, Martínez JL, Borrell YJ, Ardura A, García-Vázquez E ( 2018) Evaluating freshwater macroinvertebrates from eDNA metabarcoding: A river Nalón case study. PLoS ONE, 13, e0201741.
DOI URL |
| [22] | Ficetola GF, Miaud C, Pompanon F, Taberlet P ( 2008) Species detection using environmental DNA from water samples. Biology Letters, 4, 423-425. |
| [23] | Geerts AN, Boets P, Heede SVD, Goethals P, Heyden CVD ( 2018) A search for standardized protocols to detect alien invasive crayfish based on environmental DNA (eDNA): A lab and field evaluation. Ecological Indicators, 84, 564-572. |
| [24] |
Goldberg CS, Pilliod DS, Arkle RS, Waits LP ( 2011) Molecular detection of vertebrates in stream water: A demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE, 6, e22746.
DOI URL |
| [25] |
Goldberg CS, Sepulveda A, Ray A, Baumgardt J, Waits LP ( 2013) Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshwater Science, 32, 792-800.
DOI URL |
| [26] | Guo YH, Wang CM, Luo J, He HX ( 2009) Physa acuta found in Beijing, China. Chinese Journal of Zoology, 44(2), 127-128. (in Chinese with English abstract) |
| [ 郭云海, 王承民, 罗静, 何宏轩 ( 2009) 北京发现尖膀胱螺. 动物学杂志, 44(2), 127-128.] | |
| [27] | Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC ( 2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry, 83, 8604-8610. |
| [28] |
Jerde CL, Mahon AR, Chadderton WL, Lodge DM ( 2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conservation Letters, 4, 150-157.
DOI URL |
| [29] | Klymus KE, Marshall NT, Stepien CA ( 2017) Environmental DNA (eDNA) metabarcoding assays to detect invasive invertebrate species in the Great Lakes. PLoS ONE, 12, e0177643. |
| [30] | Kristine B, Alice E, Gilbert MTP, Carvalho GR, Simon C, Michael K, Yu DW, Mark DB ( 2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology & Evolution, 29, 358-367. |
| [31] |
Larson ER, Renshaw MA, Gantz CA, Umek J, Chandra S, Lodge DM, Egan SP ( 2017) Environmental DNA (eDNA) detects the invasive crayfishes Orconectes rusticus and Pacifastacus leniusculus in large lakes of North America. Hydrobiologia, 800, 1-13.
DOI URL |
| [32] | Li B, Ma KP ( 2010) Biological invasions: Opportunities and challenges facing Chinese ecologists in the era of translational ecology. Biodiversity Science, 18, 529-532. (in Chinese) |
| [ 李博, 马克平 ( 2010) 生物入侵: 中国学者面临的转化生态学机遇与挑战. 生物多样性, 18, 529-532.] | |
| [33] | Lowe S, Browne M, Boujdelas S, De PM ( 2000) 100 of the World’s Worst Invasive Alien Species. A selection from the Global Invasive Species Database. The IUCN Invasive Species Specialist Group, USA. |
| [34] | Majaneva M, Diserud OH, Eagle SHC, Boström E, Hajibabaei M, Ekrem T ( 2018) Environmental DNA filtration techniques affect recovered biodiversity. Scientific Reports, 8, 4682. |
| [35] | McKee AM, Spear SF, Pierson TW ( 2015) The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biological Conservation, 183, 70-76. |
| [36] | Merkes CM, Mccalla SG, Jensen NR, Gaikowski MP, Amberg JJ ( 2014) Persistence of DNA in carcasses, slime and avian feces may affect interpretation of environmental DNA data. PLoS ONE, 9, e113346. |
| [37] | Moyer GR, Díaz-Ferguson E, Hill JE, Shea C ( 2014) Assessing environmental DNA detection in controlled lentic systems. PLoS ONE, 9, e103767. |
| [38] | Piaggio AJ, Engeman RM, Hopken MW, Humphrey JS, Keacher KL, Bruce WE, Avery ML ( 2014) Detecting an elusive invasive species: A diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Molecular Ecology Resources, 14, 374-380. |
| [39] | Rees HC, Maddison BC, Middleditch DJ, Patmore James RM, Gough KC ( 2015) The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. Journal of Applied Ecology, 51, 1450-1459. |
| [40] | Renshaw MA, Olds BP, Jerde CL, McVeigh MM, Lodge DM ( 2015) The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol chloroform isoamyl alcohol DNA extraction. Molecular Ecology Resources, 15, 168-176. |
| [41] | Rosenberg DM, Resh VH ( 1993) Introduction to freshwater biomonitoring and benthic macroinvertebrates. In: Freshwater Biomonitoring and Benthic Macroinvertebrates (eds Rosenberg DM, Resh VH), pp. 1-9. Chapman & Hall, New York. |
| [42] | Shaw JLA, Clarke LJ, Wedderburn SD, Barnes TC, Weyrich LS, Cooper A ( 2016) Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biological Conservation, 197, 131-138. |
| [43] | Smith AJ, Bode RW, Kleppel GS ( 2007) A nutrient biotic index (NBI) for use with benthic macroinvertebrate communities. Ecological Indicators, 7, 371-386. |
| [44] | Strickler KM, Fremier AK, Goldberg CS ( 2015) Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation, 183, 85-92. |
| [45] |
Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH ( 2012) Environmental DNA. Molecular Ecology, 21, 1789-1793.
DOI URL |
| [46] | Takahara T, Yamanaka H, Suzuki AA, Honjo MN, Minamoto T, Yonekura R, Itayama T, Kohmatsu Y, Ito T, Kawabata ZI ( 2011) Stress response to daily temperature fluctuations in common carp, Cyprinus carpio L. Hydrobiologia, 675, 65-73. |
| [47] | Takahara T, Minamoto T , DoiDoi H (2013) Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PLoS ONE, 8, e56584. |
| [48] | Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata ZI (2012) Estimation of fish biomass using environmental DNA. PLoS ONE, 7, e35868. |
| [49] | Thomsen PF, Kielgast J, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP, Orlando L, Willerslev E ( 2012 a) Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology, 21, 2565-2573. |
| [50] | Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E ( 2012b) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE, 7, e41732. |
| [51] | Tréguier A, Paillisson J, Dejean T, Valentini A, Schlaepfer MA, Roussel J, Crispo E ( 2014) Environmental DNA surveillance for invertebrate species: Advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. Journal of Applied Ecology, 51, 871-879. |
| [52] | Tsai YL, Olson BH ( 1992) Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Applied & Environmental Microbiology, 58, 754-757. |
| [53] | Valentini A, Taberlet P, Miaud C, Civade R, Herder J, Thomsen PF, Bellemain E, Besnard A, Coissac E, Boyer F ( 2016) Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Molecular Ecology, 25, 929-942. |
| [54] |
Vamos E, Elbrecht V, Leese F ( 2017) Short COI markers for freshwater macroinvertebrate metabarcoding. Metabarcoding and Metagenomics, 1, e14625.
DOI URL |
| [55] |
Vogelstein B, Kinzler KW ( 1999) Digital PCR. Proceedings of the National Academy of Sciences, USA, 96, 9236-9241.
DOI URL |
| [56] | Wan FH ( 2009) Research on Biological Invasions in China. Science Press,Beijing. (in Chinese) |
| [ 万方浩 ( 2009) 中国生物入侵研究. 科学出版社, 北京.] | |
| [57] | Wan FH, Guo JY, Wang DH ( 2002) Alien invasive species in China: Their damages and management strategies. Biodiversity Science, 10, 119-125. (in Chinese with English abstract) |
| [ 万方浩, 郭建英, 王德辉 ( 2002) 中国外来入侵生物的危害与管理对策. 生物多样性, 10, 119-125.] | |
| [58] | Wang YJ, Li FY, Fan ZP, Cheng ZH, Zhang JQ ( 2012) The application of benthic macroinvertebrates in aquatic ecosystem health assessment. Journal of Meteorology and Environment, 28, 90-96. (in Chinese with English abstract) |
| [ 王艳杰, 李法云, 范志平, 程志辉, 张建祺 ( 2012) 大型底栖动物在水生态系统健康评价中的应用. 气象与环境学报, 28, 90-96.] | |
| [59] | Whale AS, Huggett JF, Cowen S, Speirs V, Shaw J, Ellison S, Foy CA, Scott DJ ( 2012) Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Research, 40, e82. |
| [60] | Wilcox TM, McKelvey KS, Young MK, Jane SF, Lowe WH, Whiteley AR, Schwartz MK ( 2013) Robust detection of rare species using environmental DNA: The importance of primer specificity. PLoS ONE, 8, e59520. |
| [61] | Willerslev E, Cooper A ( 2005) Ancient DNA. Proceedings of the Royal Society B: Biological Sciences, 272, 3-16. |
| [62] | Xu HG, Wu J, Liu Y, Ding H, Zhang M, Wu Y, Xi Q, Wang LL ( 2008) Biodiversity congruence and conservation strategies: A national test. BioScience, 58, 632-639. |
| [63] | Xu HG ( 2013) Introduction to the Biological Species Resources Monitor. Science Press,Beijing. (in Chinese) |
| [ 徐海根 ( 2013) 物物种资源监测概论.科学出版社, 北京.] | |
| [64] | Xu HG, Qiang S ( 2018) China’s Invasive Alien Species (revised edition).Science Press, Beijing. (in Chinese) |
| [ 徐海根, 强胜 ( 2018) 中国外来入侵生物(修订版).科学出版社, 北京.] | |
| [65] | Yang HF, Zhou WC, Qian ZX, Wang P ( 2014) An important invasive snail, Physa acuta Draparnaud 1805, found firstly in Zhejiang Province. Plant Quarantine, 28(3), 63-65. (in Chinese with English abstract) |
| [ 杨海芳, 周卫川, 钱周兴, 王沛 ( 2014) 浙江发现重要外来入侵物种——尖膀胱螺. 植物检疫, 28(3), 63-65.] | |
| [66] | Yeom DH, Adams SM ( 2007) Assessing effects of stress across levels of biological organization using an aquatic ecosystem health index. Ecotoxicology & Environmental Safety, 67, 286-295. |
| [1] | 贺加贝, 柯可, 孙海明, 胡丽萍, 赵晓伟, 王文豪, 赵强. 基于DNA宏条形码技术分析香螺食性[J]. 生物多样性, 2025, 33(1): 24403-. |
| [2] | 耿宜佳, 田瑜, 李俊生, 李子圆, 潘玉雪. 《生物多样性公约》框架下外来入侵物种管控的全球进展、挑战和展望[J]. 生物多样性, 2024, 32(11): 24275-. |
| [3] | 董志远, 陈琳琳, 张乃鹏, 陈莉, 孙德斌, 倪艳梅, 李宝泉. 基于环境DNA宏条形码技术研究黄河三角洲典型潮沟系统鱼类多样性及其对水文连通性的响应[J]. 生物多样性, 2023, 31(7): 23073-. |
| [4] | 湛振杰, 张超, 陈敏豪, 王嘉栋, 富爱华, 范雨薇, 栾晓峰. 基于DNA宏条形码技术的大兴安岭北部欧亚水獭冬季食性分析[J]. 生物多样性, 2023, 31(6): 22586-. |
| [5] | 彭步青, 陶玲, 李靖, 范荣辉, 陈顺德, 付长坤, 王琼, 唐刻意. 基于DNA宏条形码研究四川老君山国家级自然保护区6种同域共存小型哺乳动物的食性[J]. 生物多样性, 2023, 31(4): 22474-. |
| [6] | 沈梅, 郭宁宁, 罗遵兰, 郭晓晨, 孙光, 肖能文. 基于eDNA metabarcoding探究北京市主要河流鱼类分布及影响因素[J]. 生物多样性, 2022, 30(7): 22240-. |
| [7] | 李晗溪, 黄雪娜, 李世国, 战爱斌. 基于环境DNA-宏条形码技术的水生生态系统入侵生物的早期监测与预警[J]. 生物多样性, 2019, 27(5): 491-504. |
| [8] | 刘硕然, 杨道德, 李先福, 谭路, 孙军, 和晓阳, 杨文书, 任国鹏, Davide Fornacca, 蔡庆华, 肖文. 滇西北高山微水体与溪流生境底栖动物多样性和环境特征[J]. 生物多样性, 2019, 27(12): 1298-1308. |
| [9] | 郎丹丹, 唐敏, 周欣. 传粉网络构建的定性定量分子研究: 应用与展望[J]. 生物多样性, 2018, 26(5): 445-456. |
| [10] | 陈良燕, 徐海根. 澳大利亚外来入侵物种管理策略及对我国的借鉴意义[J]. 生物多样性, 2001, 09(4): 466-471. |
| [11] | 刘焕章, 陈宜瑜. 淡水生态系统中的TOP-DOWN效应与生物多样性保护[J]. 生物多样性, 1996, 04(2): 109-113. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn