



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (2): 24434. DOI: 10.17520/biods.2024434 cstr: 32101.14.biods.2024434
• Data Papers • Previous Articles Next Articles
Lin Zhen1( ), Xiang Jiabao1(
), Xiang Jiabao1( ), Cai Hejiayi1(
), Cai Hejiayi1( ), Gao Bei1(
), Gao Bei1( ), Yang Jintao1(
), Yang Jintao1( ), Li Junyi1(
), Li Junyi1( ), Zhou Qingsong2(
), Zhou Qingsong2( ), Huang Xiaolei1,*(
), Huang Xiaolei1,*( )(
)( ), Deng Jun1,*(
), Deng Jun1,*( )(
)( )
)
Received:2024-10-01
Accepted:2024-12-10
Online:2025-02-20
Published:2025-03-14
Contact:
*E-mail: huangxl@fafu.edu.cn;
tiancai1282008@126.com
Supported by:Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species[J]. Biodiv Sci, 2025, 33(2): 24434.
| 物种 Species | 科 Family | 属 Genus | SRA检索号 SRA accession number | GenBank登录号 GenBank accession number | 采集地点 Collection locality | 序列长度 Sequence length (bp) |
|---|---|---|---|---|---|---|
| Maevius indecorus | 锚蝽科 Hyocephalidae | Maevius | SRR6413622 | PQ191211 | 澳大利亚昆士兰 Queensland, Australia | 17,047 |
| Limnogonus cereiventris | 水黾科 Gerridae | 泽背黾蝽属 Limnogonus | SRR9110347 | PQ389959 | 喀麦隆布埃亚 Buea, Cameroon | 15,474 |
| 尖长蝽 Oxycarenus laetus | 尖长蝽科 Oxycarenidae | 尖长蝽属 Oxycarenus | SRR11024516 | PQ191210 | 印度珀丁达 Bhatinda, India | 15,921 |
| Hormaphis cornu | 蚜科 Aphididae | 扁蚜属 Hormaphis | SRR11363133 | PQ336989 | 美国新泽西州 New Jersey, USA | 16,198 |
| Melanaphis donacis | 蚜科 Aphididae | 色蚜属 Melanaphis | SRR26205096 | PQ323560 | 美国加利福尼亚州 California, USA | 17,116 |
| 香蕉交脉蚜 Pentalonia nigronervosa | 蚜科 Aphididae | 交脉蚜属 Pentalonia | SRR11607999 | PQ327933 | 肯尼亚内罗毕 Nairobi, Kenya | 15,115 |
| 石榴囊毡蚧 Acanthococcus lagerstroemiae* | 毡蚧科 Eriococcidae | 毡蚧属 Acanthococcus | SRR18768944 | PQ390451 | 中国石家庄 Shijiazhuang, China | 14,219 |
Table 1 The sample information on seven species of Hemiptera in this study
| 物种 Species | 科 Family | 属 Genus | SRA检索号 SRA accession number | GenBank登录号 GenBank accession number | 采集地点 Collection locality | 序列长度 Sequence length (bp) |
|---|---|---|---|---|---|---|
| Maevius indecorus | 锚蝽科 Hyocephalidae | Maevius | SRR6413622 | PQ191211 | 澳大利亚昆士兰 Queensland, Australia | 17,047 |
| Limnogonus cereiventris | 水黾科 Gerridae | 泽背黾蝽属 Limnogonus | SRR9110347 | PQ389959 | 喀麦隆布埃亚 Buea, Cameroon | 15,474 |
| 尖长蝽 Oxycarenus laetus | 尖长蝽科 Oxycarenidae | 尖长蝽属 Oxycarenus | SRR11024516 | PQ191210 | 印度珀丁达 Bhatinda, India | 15,921 |
| Hormaphis cornu | 蚜科 Aphididae | 扁蚜属 Hormaphis | SRR11363133 | PQ336989 | 美国新泽西州 New Jersey, USA | 16,198 |
| Melanaphis donacis | 蚜科 Aphididae | 色蚜属 Melanaphis | SRR26205096 | PQ323560 | 美国加利福尼亚州 California, USA | 17,116 |
| 香蕉交脉蚜 Pentalonia nigronervosa | 蚜科 Aphididae | 交脉蚜属 Pentalonia | SRR11607999 | PQ327933 | 肯尼亚内罗毕 Nairobi, Kenya | 15,115 |
| 石榴囊毡蚧 Acanthococcus lagerstroemiae* | 毡蚧科 Eriococcidae | 毡蚧属 Acanthococcus | SRR18768944 | PQ390451 | 中国石家庄 Shijiazhuang, China | 14,219 |
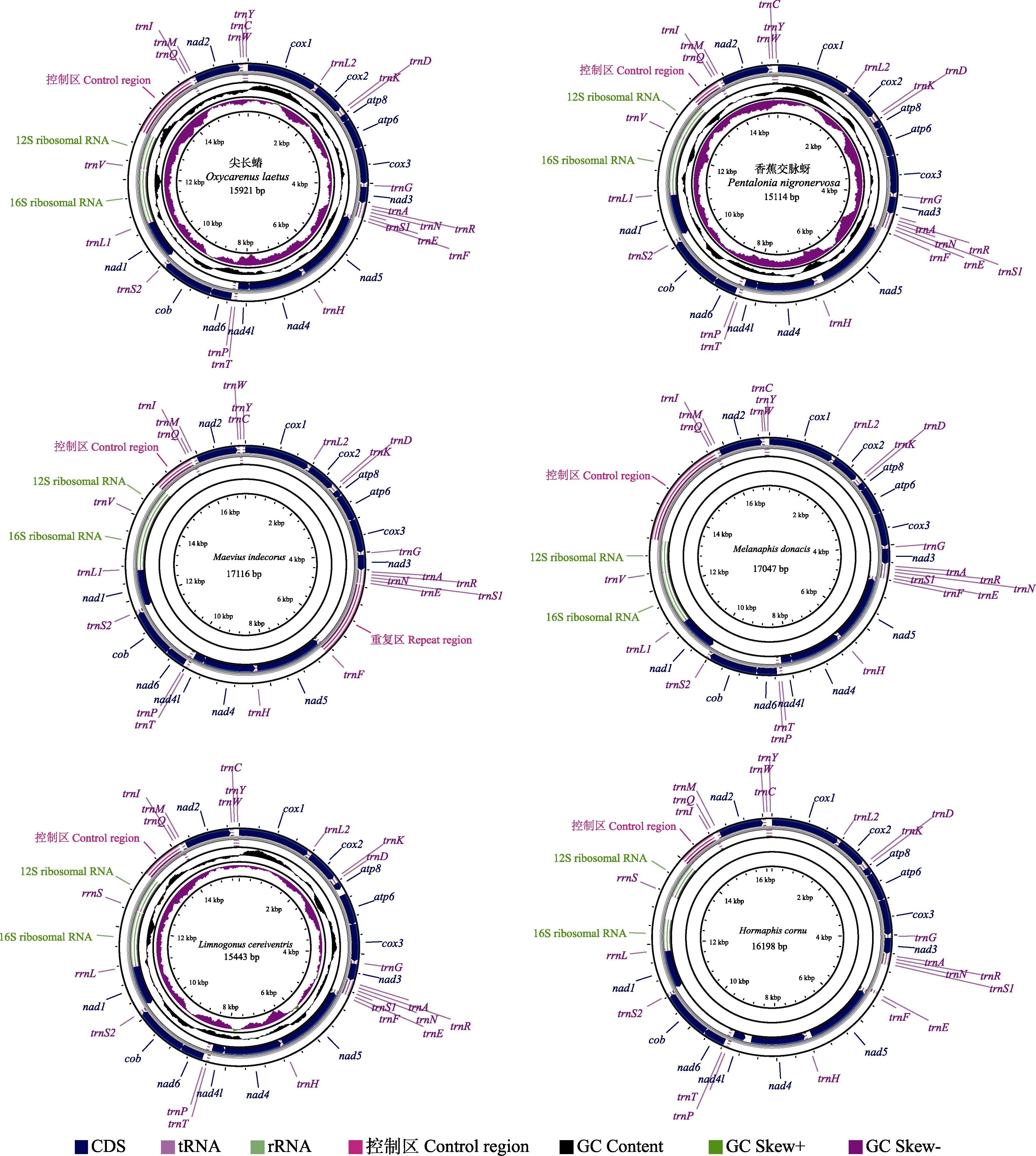
Fig. 1 Mitogenome circular maps of the Maevius indecorus, Limnogonus cereiventris, Oxycarenus laetus, Hormaphis cornu, Melanaphis donacis, and Pentalonia nigronervosa. The genes that are labeled on the outside circle indicate that coding sequences are on the major (J) strand, while others are on the minor (N) strand. The tRNA genes are labeled with the amino acid abbreviations. The incomplete mitochondrial genome has not been mapped.
| [1] | Bánki O, Roskov Y, Döring M, Ower G, Hernández Robles DR, Plata Corredor CA, Stjernegaard Jeppesen T, Örn A, Pape T, Hobern D, Garnett S, Little H, DeWalt RE, Ma K, Miller J, Orrell T, Aalbu R, Abbott J, Aedo C, et al. (2024) Catalogue of Life (Version 2024-08-29). Catalogue of Life, Amsterdam, Netherlands. https://doi.org/10.48580/dgdwl. |
| [2] | Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69, 313-319. |
| [3] |
Burger G, Gray MW, Franz Lang B (2003) Mitochondrial genomes: Anything goes. Trends in Genetics, 19, 709-716.
PMID |
| [4] | Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45, e18. |
| [5] | Grant JR, Stothard P (2008) The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Research, 36, W181-W184. |
| [6] |
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874.
DOI PMID |
| [7] | Li DH, Liu CM, Luo RB, Sadakane K, Lam TW (2015) MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 31, 1674-1676. |
| [8] | Lu CC, Huang XL, Deng J (2020) The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. International Journal of Biological Macromolecules, 158, 854-864. |
| [9] | Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng QD, Wortman J, Young SK, Earl AM (2014) Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE, 9, e112963. |
| [10] |
Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348-355.
DOI PMID |
| [1] | Jing Gan Xiangxu Liu Xueming Lu Xing Yue. China's Large Cities in Global Biodiversity Hotspots: Conservation Policies and Optimization Directions [J]. Biodiv Sci, 2025, 33(5): 24529-. |
| [2] | Zixuan Zeng Rui Yang Yue Huang Luyao Chen. Characteristics of bird diversity and environmental relationships in Tsinghua University campus [J]. Biodiv Sci, 2025, 33(5): 24373-. |
| [3] | Mingyue Zang, Li Liu, Yue Ma, Xu Xu, Feilong Hu, Xiaoqiang Lu, Jiaqi Li, Cigang Yu, Yan Liu. China’s urban biodiversity conservation under the Kunming-Montreal Global Biodiversity Framework [J]. Biodiv Sci, 2025, 33(5): 24482-. |
| [4] | Xiaoyu Zhu, Chenhao Wang, Zhongjun Wang, Yujun Zhang. Research progress and prospect of urban green space biodiversity [J]. Biodiv Sci, 2025, 33(5): 25027-. |
| [5] | Lin Yuan, Siqi Wang, Jingxuan Hou. “Leaving space for wildness” in metropolitan region: Trends and prospects [J]. Biodiv Sci, 2025, 33(5): 24481-. |
| [6] | Min Hu, Binbin Li, Coraline Goron. Green is not enough: A management framework for urban biodiversity-friendly parks [J]. Biodiv Sci, 2025, 33(5): 24483-. |
| [7] | Xin Wang, Femgyu Bao. Analysis of the ecological restoration effect of South Dianchi National Wetland Park based on the enhancement of bird diversity [J]. Biodiv Sci, 2025, 33(5): 24531-. |
| [8] | Yue Ming, Peiyao Hao, Lingqian Tan, Xi Zheng. A study on urban biodiversity conservation and enhancement in china based on the concept of green and high-quality development of cities [J]. Biodiv Sci, 2025, 33(5): 24524-. |
| [9] | Gan Xie, Jing Xuan, Qidi Fu, Ze Wei, Kai Xue, Hairui Luo, Jixi Gao, Min Li. Establishing an intelligent identification model for unmanned aerial vehicle surveys of grassland plant diversity [J]. Biodiv Sci, 2025, 33(4): 24236-. |
| [10] | Xiaolin Chu, Quanguo Zhang. A review of experimental evidence for the evolutionary speed hypothesis [J]. Biodiv Sci, 2025, 33(4): 25019-. |
| [11] | Zhiyu Liu, Xin Ji, Guohui Sui, Ding Yang, Xuankun Li. Invertebrate diversity in buffalo grass and weedy lawns at Beijing Capital International Airport [J]. Biodiv Sci, 2025, 33(4): 24456-. |
| [12] | Xiaoqiang Lu, Shanshan Dong, Yue Ma, Xu Xu, Feng Qiu, Mingyue Zang, Yaqiong Wan, Luanxin Li, Cigang Yu, Yan Liu. Current status, challenges, and prospects of frontier technologies in biodiversity conservation applications [J]. Biodiv Sci, 2025, 33(4): 24440-. |
| [13] | Qiaoyi Nong, Jun Cao, Wenda Cheng, Yanqiong Peng. Comparative study of monitoring methods for Apoidea resources and diversity [J]. Biodiv Sci, 2025, 33(4): 25057-. |
| [14] | Guo Yutong, Li Sucui, Wang Zhi, Xie Yan, Yang Xue, Zhou Guangjin, You Chunhe, Zhu Saning, Gao Jixi. Coverage and distribution of national key protected wild species in China’s nature reserves [J]. Biodiv Sci, 2025, 33(3): 24423-. |
| [15] | Zhao Weiyang, Wang Wei, Ma Bingran. Advances and prospects in research on other effective area-based conservation measures (OECMs) [J]. Biodiv Sci, 2025, 33(3): 24525-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()