



Biodiv Sci ›› 2019, Vol. 27 ›› Issue (11): 1221-1227. DOI: 10.17520/biods.2019236 cstr: 32101.14.biods.2019236
• Original Papers • Previous Articles Next Articles
Hao Wang1,2#,Rui Zhang1#,Jiao Zhang1,Hui Shen1,Xiling Dai2,Yuehong Yan1,*( )
)
Received:2019-07-25
Accepted:2019-12-17
Online:2019-11-20
Published:2020-01-17
Contact:
Yan Yuehong
Hao Wang, Rui Zhang, Jiao Zhang, Hui Shen, Xiling Dai, Yuehong Yan. De novo transcriptome assembly reveals the whole genome duplication events of Didymochlaena trancatula[J]. Biodiv Sci, 2019, 27(11): 1221-1227.
| 转录本 Transcript | 单基因簇 Unigene | |
|---|---|---|
| 序列数目 Sequence number | 77,709 | 58,871 |
| 平均长度 Mean length (bp) | 986 | 836 |
| 最大长度 Max. length (bp) | 17,374 | 17,374 |
| 最小长度 Min. length (bp) | 201 | 201 |
| N50 | 1,734 | 1,546 |
Table 1 Summary of de novo assembly for Didymochlaena trancatula transcriptome
| 转录本 Transcript | 单基因簇 Unigene | |
|---|---|---|
| 序列数目 Sequence number | 77,709 | 58,871 |
| 平均长度 Mean length (bp) | 986 | 836 |
| 最大长度 Max. length (bp) | 17,374 | 17,374 |
| 最小长度 Min. length (bp) | 201 | 201 |
| N50 | 1,734 | 1,546 |
| 加倍数目 No. of duplications | 组件数目 No. of components | 贝叶斯信息标准 Bayesian information criterion | 中值 Median | 方差 Variance | 比例 Proportion |
|---|---|---|---|---|---|
| 1,951 | 7 | 5,012.931 | 0.1211 | 0.0002 | 0.0953 |
| 1,951 | 7 | 5,012.931 | 0.1796 | 0.0015 | 0.0897 |
| 1,951 | 7 | 5,012.931 | 0.3518 | 0.0134 | 0.0584 |
| 1,951 | 7 | 5,012.931 | 1.3359 | 0.1921 | 0.3296 |
| 1,951 | 7 | 5,012.931 | 2.0329 | 0.1872 | 0.2107 |
| 1,951 | 7 | 5,012.931 | 3.2182 | 0.5590 | 0.1958 |
| 1,951 | 7 | 5,012.931 | 4.7933 | 0.0105 | 0.0205 |
Table 2 Ks results of Didymochlaena trancatula based on mixture modeling
| 加倍数目 No. of duplications | 组件数目 No. of components | 贝叶斯信息标准 Bayesian information criterion | 中值 Median | 方差 Variance | 比例 Proportion |
|---|---|---|---|---|---|
| 1,951 | 7 | 5,012.931 | 0.1211 | 0.0002 | 0.0953 |
| 1,951 | 7 | 5,012.931 | 0.1796 | 0.0015 | 0.0897 |
| 1,951 | 7 | 5,012.931 | 0.3518 | 0.0134 | 0.0584 |
| 1,951 | 7 | 5,012.931 | 1.3359 | 0.1921 | 0.3296 |
| 1,951 | 7 | 5,012.931 | 2.0329 | 0.1872 | 0.2107 |
| 1,951 | 7 | 5,012.931 | 3.2182 | 0.5590 | 0.1958 |
| 1,951 | 7 | 5,012.931 | 4.7933 | 0.0105 | 0.0205 |
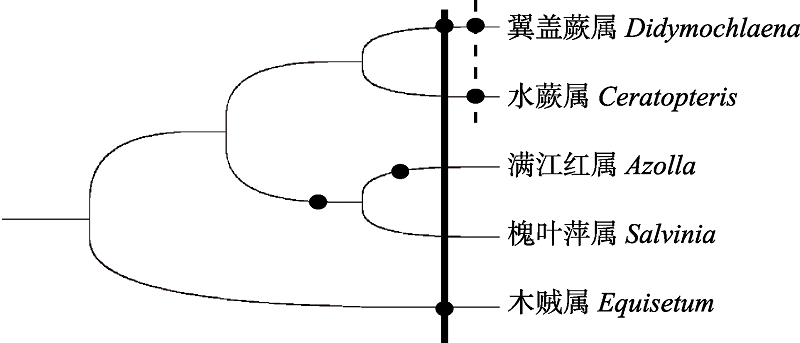
Fig. 3 WGDs occurrence in ferns from previous reports. Circle represents WGDs. The solid line denotes the age of WGD at (about 90 Mya) Didymochlaena trancatula and Equisetum giganteum, the dash line indicates the age of WGD both detected in Didymochlaena trancatula and Ceretopteris thalictroides (about 50 Mya).
| 1 | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G ( 2000) Gene ontology: Tool for the unification of biology. Nature Genetic, 25, 25-29. |
| 2 | Athanasios T, Joseph RE, Ronald WD ( 2000) Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature, 408, 816-820. |
| 3 | Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B, Legrand L, Gill N, Kane NC, Bowers JE, Hubner S, Bellec A, Bérard A, Bergès H, Blanchet N, Boniface MC, Brunel D, Catrice O, Chaidir N, Claudel C, Donnadieu C, Faraut T, Fievet G, Helmstetter N, King M, Knapp SJ, Lai Z, Le Paslier MC, Lippi Y, Lorenzon L, Mandel JR, Marage G, Marchand G, Marquand E, Bret-Mestries E, Morien E, Nambeesan S, Nguyen T, Pegot-Espagnet P, Pouilly N, Raftis F, Sallet E, Schiex T, Thomas J, Vandecasteele C, Varès D, Vear F, Vautrin S, Crespi M, Mangin B, Burke JM, Salse J, Muños S, Vincourt P, Rieseberg LH, Langlade NB ( 2017) The sunflower genome provides insights into oil metabolism, flowering and asterid evolution. Nature, 546, 148-152. |
| 4 | Barker MS ( 2009) Evolutionary genomic analyses of ferns reveal that high chromosome numbers are a product of high retention and fewer rounds of polyploidy relative to angiosperms. American Fern Journal, 99, 136-141. |
| 5 | Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA ( 2015) On the relative abundance of auto- and allopolyploids. New Phytologist, 210, 391-398. |
| 6 | Blanc G, Wolfe KH ( 2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell, 16, 1667-1678. |
| 7 | Bowers JE, Chapman BA, Rong J, Paterson AH ( 2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature, 422, 433-438. |
| 8 | Clark JW, Puttick MN, Donoghue PCJ ( 2019) Origin of horsetails and the role of whole-genome duplication in plant macroevolution. Proceedings of the Royal Society B: Biological Sciences, 286, 20191662. |
| 9 | Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, Albert VA, Ma H, dePamphilis CW ( 2006) Widespread genome duplications throughout the history of flowering plants. Genome Research, 16, 738-749. |
| 10 | De Bodt S, Maere S, Van de Peer Y ( 2005) Genome duplication and the origin of angiosperms. Trends in Ecology & Evolution, 20, 591-597. |
| 11 | Dyer RJ, Pellicer J, Savolainen V, Leitch IJ, Schneider H ( 2013) Genome size expansion and the relationship between nuclear DNA content and spore size in the Asplenium monanthes fern complex (Aspleniaceae). BMC Plant Biology, 13, 219. |
| 12 | Edgar RC ( 2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792-1797. |
| 13 | Enright AJ, Van Dongen S, Ouzounis CA ( 2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Research, 30, 1575-1584. |
| 14 | Fawcett JA, Maere S, Van de Peer Y ( 2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proceedings of the National Academy of Sciences, USA, 106, 5737-5742. |
| 15 | Fraley C, Raftery AE ( 2003) Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST. Journal of Classification, 20, 263-286. |
| 16 | Freeling M ( 2009) Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annual Review of Plant Biology, 60, 433-453. |
| 17 | Goldman N, Yang Z ( 1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution, 11, 725-736. |
| 18 | Guerra D, Crosatti C, Khoshro HH, Mastrangelo AM, Mica E, Mazzucotelli E ( 2015) Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Frontier in Plant Science, 6, 57. |
| 19 | Hegarty MJ, Hiscock SJ ( 2008) Genomic clues to the evolutionary success of polyploid plants. Current Biology, 18, 435-444. |
| 20 | Huang CH, Qi X, Chen D, Qi J, Ma H ( 2019) Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. Journal of Integrative Plant Biology, doi: 10.1111/jipb.12877. |
| 21 | Huang CH, Zhang C, Liu M, Hu Y, Gao T, Qi J, Ma H ( 2016) Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Molecular Biology and Evolution, 33, 2820-2835. |
| 22 | Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW ( 2011) Ancestral polyploidy in seed plants and angiosperms. Nature, 473, 97-100. |
| 23 | Kimura M ( 1977) Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature, 267, 275-276. |
| 24 | Li FW, Brouwer P, Carretero-Paulet L, Cheng S, de Vries J, Delaux PM, Eily A, Koppers N, Kuo LY, Li Z, Simenc M, Small I, Wafula E, Angarita S, Barker MS, Bräutigam A, dePamphilis C, Gould S, Hosmani PS, Huang YM, Huettel B, Kato Y, Liu X, Maere S, McDowell R, Mueller LA, Nierop KGJ, Rensing SA, Robison T, Rothfels CJ, Sigel EM, Song Y, Timilsena PR, Van de Peer Y, Wang H, Wilhelmsson PKI, Wolf PG, Xu X, Der JP, Schluepmann H, Wong GK, Pryer KM ( 2018) Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nature Plants, 4, 460-472. |
| 25 | Lynch M, Conery JS ( 2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151-1155. |
| 26 | Muñoz A, Castellano MM ( 2012) Regulation of translation initiation under abiotic stress conditions in plants: It is a conserved or not so conserved process among eukaryotes? Comparative and Functional Genomics, 2012,406357. |
| 27 | Niu QK, Liang Y, Zhou JJ, Dou XY, Gao SC, Chen LQ, Zhang XQ, Ye D ( 2013) Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in Arabidopsis. Molecular Plant, 6, 1091-1098. |
| 28 | One Thousand Plant Transcriptomes Initiative ( 2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574, 679-685. |
| 29 | PPG I ( 2016) A community-derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution, 54, 563-603. |
| 30 | Rothfels CJ, Li FW, Sigel EM, Huiet L, Larsson A, Burge DO, Ruhsam M, Deyholos M, Soltis DE, Stewart CN Jr, Shaw SW, Pokorny L, Chen T, dePamphilis C, DeGironimo L, Chen L, Wei X, Sun X, Korall P, Stevenson DW, Graham SW, Wong GK, Pryer KM ( 2015) The evolutionary history of ferns inferred from 25 low-copy nuclear genes. American Journal of Botany, 102, 1089-1107. |
| 31 | Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC ( 2004) Mining EST databases to resolve evolutionary events in major crop species. Genome, 47, 868-876. |
| 32 | Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallon S, Lupia R ( 2004) Ferns diversified in the shadow of angiosperms. Nature, 428, 553-557. |
| 33 | Semon M, Wolfe KH ( 2007) Reciprocal gene loss between tetraodon and zebrafish after whole genome duplication in their ancestor. Trends in Genetics, 23, 108-112. |
| 34 | Smith AR, Cranfill RB ( 2002) Intrafamilial relationships of the thelypteroid ferns (Thelypteridaceae). American Fern Journal, 92, 131-149. |
| 35 | Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, depamphilis CW, Wall PK, Soltis PS ( 2009) Polyploidy and angiosperm diversification. American Journal of Botany, 96, 336-348. |
| 36 | Soltis PS, Marchant DB, Van de Peer Y, Soltis DE ( 2015) Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development, 35, 119-125. |
| 37 | Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z ( 2017) agriGO v2.0: A GO analysis toolkit for the agricultural community. Nucleic Acids Research, 45, 122-129. |
| 38 | Tsutsumi C, Kato M ( 2005) Molecular phylogenetic study on Davalliaceae. Fern Gazatte, 17, 147-162. |
| 39 | Tsutsumi C, Kato M ( 2006) Evolution of epiphytes in Davalliaceae and related ferns. Botanical Journal of the Linnean Society, 151, 495-510. |
| 40 | Vajda V, Raine JI, Hollis CJ ( 2001) Indication of global deforestation at the Cretaceous-Tertiary boundary by New Zealand fern spike. Science, 294, 1700-1702. |
| 41 | Van de Peer Y, Maere S, Meyer A ( 2009) The evolutionary significance of ancient genome duplications. Nature Reviews Genetics, 10, 725-732. |
| 42 | Vanneste K, Baele G, Maere S, Van de Peer Y ( 2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Research, 24, 1334-1347. |
| 43 | Vanneste K, Sterck L, Myburg AA, Van de Peer Y, Mizrachi E ( 2015) Horsetails are ancient polyploids: Evidence from Equisetum giganteum. The Plant Cell, 27, 1567-1578. |
| 44 | Vanneste K, Van de Peer Y, Maere S ( 2013) Inference of genome duplications from age distributions revisited. Molecular Biology and Evolution, 30, 177-190. |
| 45 | Wang JT, Li JT, Zhang XF, Sun XW ( 2012) Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genomics, 13, 96. |
| 46 | Wolfe KH, Shields DC ( 1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708-713. |
| 47 | Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH ( 2009) The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA, 106, 13875-13879. |
| 48 | Xiang YZ, Huang CH, Yi H, Wen J, Li S, Yi T, Chen H, Xiang J, Hong M ( 2017) Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Molecular Biology and Evolution, 34, 262-281. |
| 49 | Yang Z ( 2007) PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24, 1586-1591. |
| 50 | Zhang LB, Zhang L ( 2015) Didymochlaenaceae: A new fern family of eupolypods I (Polypodiales). Taxon, 64, 27-38. |
| 51 | Zhang R, Wang FG, Zhang J, Shang H, Liu L, Wang H, Zhao GH, Shen H, Yan YH ( 2019) Dating whole genome duplication in Ceratopteris thalictroides and potential adaptive values of retained gene duplicates. International Journal of Molecular Science, 20, 1926. |
| [1] | Min Hu, Binbin Li, Coraline Goron. Green is not enough: A management framework for urban biodiversity-friendly parks [J]. Biodiv Sci, 2025, 33(5): 24483-. |
| [2] | Nan Chen, Quan-Guo Zhang. The experimental evolution approach [J]. Biodiv Sci, 2024, 32(9): 24171-. |
| [3] | Yihui Jiang, Yue Liu, Xu Zeng, Zheying Lin, Nan Wang, Jihao Peng, Ling Cao, Cong Zeng. Fish diversity and connectivity in six national marine protected areas in the East China Sea [J]. Biodiv Sci, 2024, 32(6): 24128-. |
| [4] | Zhirong Feng, Youcheng Chen, Yanqiong Peng, Li Li, Bo Wang. Ecological network analysis: From metacommunity to metanetwork [J]. Biodiv Sci, 2023, 31(8): 23171-. |
| [5] | Tianao Chen, Xiang Li. Identifying the management system for national parks in China [J]. Biodiv Sci, 2023, 31(3): 22485-. |
| [6] | Zhijing Xie, Xiangyu Liu, Xiaoming Sun, Jiliang Liu, Zhanfeng Liu, Xiaoke Zhang, Jun Chen, Xiaodong Yang, Bo Zhu, Xin Ke, Donghui Wu. Advances and prospects of the thematic monitoring network of soil fauna diversity in China [J]. Biodiv Sci, 2023, 31(12): 23365-. |
| [7] | Shumei Zhang, Wei Li, Dingnan Li. Inventory of species diversity of Liaoning higher plants [J]. Biodiv Sci, 2022, 30(6): 22038-. |
| [8] | Qing Yang, Peng Zhang, Ruizhi An, Nanqian Qiao, Zhen Da, Sang Ba. Spatial and temporal distribution patterns and driving mechanisms of ciliate communities in the midstream and downstream reaches of the Lhasa River [J]. Biodiv Sci, 2022, 30(6): 22012-. |
| [9] | Wei Wang, Yue Zhou, Yu Tian, Junsheng Li. Biodiversity conservation research in protected areas: A review [J]. Biodiv Sci, 2022, 30(10): 22459-. |
| [10] | Ting Wang, Zengqiang Xia, Jiangping Shu, Jiao Zhang, Meina Wang, Jianbing Chen, Kanglin Wang, Jianying Xiang, Yuehong Yan. Dating whole-genome duplication reveals the evolutionary retardation of Angiopteris [J]. Biodiv Sci, 2021, 29(6): 722-734. |
| [11] | Liang Hu. Species diversity and geographical distribution of marine, benthic, shell-bearing mollusks on the coast and adjacent area of Pingtan Island, Fujian Province [J]. Biodiv Sci, 2021, 29(10): 1403-1410. |
| [12] | Lü Zhongmei. Systematic legislation for nature conservation with national parks as the main body [J]. Biodiv Sci, 2019, 27(2): 128-136. |
| [13] | Xinyu Du, Jinmei Lu, Dezhu Li. Advances in the evolution of plastid genome structure in lycophytes and ferns [J]. Biodiv Sci, 2019, 27(11): 1172-1183. |
| [14] | Guohua Zhao, Ying Wang, Hui Shang, Xile Zhou, Aihua Wang, Yufeng Li, Hui Wang, Baodong Liu, Yuehong Yan. Ancestral state reconstruction reveals the diversity and evolution of spore ornamentation in Adiantum (Pteridaceae) [J]. Biodiv Sci, 2019, 27(11): 1228-1235. |
| [15] | Zhen Zhan, Jianfeng Zhang, Jianguo Cao, Xiling Dai. Development of archegonium and oogenesis of the fern Macrothelypteris torresiana [J]. Biodiv Sci, 2019, 27(11): 1236-1244. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn