



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (1): 76-82. DOI: 10.3724/SP.J.1003.2010.076 cstr: 32101.14.SP.J.1003.2010.076
• Original Papers • Previous Articles Next Articles
Yu Zheng1,3, Bo Gao4, Lifu Sun1,2, Yanhong Bing1, Kequan Pei1,*( )
)
Received:2009-10-14
Accepted:2009-11-27
Online:2010-01-20
Published:2010-01-20
Contact:
Kequan Pei
Yu Zheng, Bo Gao, Lifu Sun, Yanhong Bing, Kequan Pei. Diversity of fungi associated with Rhododendron argyrophyllum and R. floribundum hair roots in Sichuan, China[J]. Biodiv Sci, 2010, 18(1): 76-82.
| 克隆 Clone | 序列号 GenBank accession no. | BLAST最接近的比对 Closest BLAST match | 重叠区 Overlap (bp) | 相似度 Identity (%) | 克隆数目 Clone No. | ||
|---|---|---|---|---|---|---|---|
| RA | RF | ||||||
| 子囊菌纲 Ascomycota | |||||||
| 061229 | GU256941 | Alternaria alternate (DQ023279) | 608 | 100 | 1 | 3 | |
| 0612271 | GU256942 | — | — | — | 1 | 0 | |
| 061266 | GU256943 | Uncultured Geoglossaceae clone Y43 (DQ273321) | 565 | 90.2 | 8 | 6 | |
| 061626 | GU256944 | Uncultured Rhizoscyphus ericae complex clone K10-16 (AY394687) | 591 | 93.3 | 1 | 5 | |
| 061206 | GU256945 | Uncultured Rhizoscyphus ericae complex clone K10-16 (AY394687) | 550 | 92.5 | 7 | 12 | |
| 061619 | GU256946 | Rhizoscyphus ericae aggregate (AM084704) | 573 | 94.5 | 0 | 2 | |
| 061239 | GU256947 | Rhizoscyphus ericae aggregate (AY394684) | 550 | 87.1 | 3 | 0 | |
| 061286 | GU256948 | Oidiodendron maius strain UAMH 8921 (AF062800) | 527 | 99.1 | 9 | 0 | |
| 061210 | GU256949 | Cryptosporiopsis ericae voucher UAMH 10419 (AY853167) | 599 | 88.1 | 1 | 0 | |
| 061235 | GU256950 | Phialea strobilina strain CBS 643.85 (EF596821) | 571 | 88.3 | 3 | 0 | |
| 061214 | GU256951 | Salal root associated fungus UBCtra1011.13 (AF300726) | 367 | 99.2 | 1 | 0 | |
| 061655 | GU256952 | Mycorrhizal ascomycete of Rhododendron type 3 (AB089663) | 522 | 93.3 | 0 | 4 | |
| 061223 | GU256953 | Mycorrhizal ascomycete of Rhododendron type 2 (AB089660) | 521 | 95.4 | 1 | 0 | |
| 061245 | GU256954 | Mycorrhizal ascomycete of Rhododendron type 2 (AB089660) | 530 | 80.8 | 4 | 0 | |
| 061289 | GU256955 | Penicillium griseofulvum (AB369901) | 577 | 99.6 | 12 | 0 | |
| 061642 | GU256956 | Penicillium sp. 5/97-5 (AJ279476) | 615 | 99.1 | 2 | 2 | |
| 061211 | GU256957 | Paecilomyces sp. JCM 12545 (AB217857) | 632 | 100 | 1 | 0 | |
| 担子菌纲 Basidiomyota | |||||||
| 061221 | GU256958 | Erythrobasidium hasegawianum strain CBS 8253 (AF444522) | 617 | 85.5 | 1 | 0 | |
| 061284 | GU256959 | Erythrobasidium hasegawianum strain CBS 8253 (AF444522) | 635 | 81.6 | 1 | 0 | |
| 061230 | GU256960 | Sporobolomyces yunnanensis (AB030353) | 601 | 99.1 | 10 | 0 | |
| 061277 | GU256961 | Uncultured ectomycorrhiza (Basidiomycota) (AY641466) | 701 | 91.1 | 4 | 0 | |
| 061201 | GU256962 | Cryptococcus terricola strain CBS 6435 (AF444377) | 645 | 100 | 2 | 0 | |
| 061241 | GU256963 | Cryptococcus sp. BF73 (AM901689) | 631 | 98.3 | 1 | 0 | |
| 061624 | GU256964 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 542 | 79.4 | 0 | 14 | |
| 061265 | GU256965 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 542 | 82.4 | 2 | 0 | |
| 061647 | GU256966 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 536 | 89.7 | 0 | 3 | |
| 061692 | GU256967 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 542 | 80.8 | 0 | 1 | |
| 061632 | GU256968 | Uncultured cf. Tricholoma (AY097046) | 670 | 98.5 | 0 | 2 | |
| 061628 | GU256969 | Uncultured ectomycorrhiza (Tricholomataceae) (AM113458) | 645 | 85.4 | 1 | 20 | |
| 061610 | GU256970 | Tricholoma sp. Umpqua (AF377204) | 725 | 82.6 | 0 | 1 | |
| 061207 | GU256971 | Uncultured mycorrhiza (Sebacinaceae) 4078 (AY634132) | 664 | 84.6 | 5 | 0 | |
| 061220 | GU256972 | Uncultured mycorrhiza (Sebacinales) voucher 04A16 (DQ352049) | 494 | 88.5 | 1 | 0 | |
| 0616101 | GU256973 | Uncultured Sebacinales clone R33 (EF030897) | 412 | 93.0 | 0 | 1 | |
| 061203 | GU256974 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 674 | 92.8 | 2 | 20 | |
| 061209 | GU256975 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 674 | 91.6 | 2 | 0 | |
| 061606 | GU256976 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 671 | 91.6 | 0 | 2 | |
| 061262 | GU256977 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 673 | 90.6 | 4 | 0 | |
| 061607 | GU256978 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 689 | 96.3 | 0 | 1 | |
| 061267 | GU256979 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 679 | 92.1 | 1 | 0 | |
| 061668 | GU256980 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 669 | 91.2 | 0 | 2 | |
| 061675 | GU256981 | Gymnopus austrosemihirtipes AWW65 (AY263422) | 796 | 95.3 | 0 | 4 | |
Table 1 The number of each ITS taxa clones from root systems of Rhododendron argyrophyllum (RA) and R. floribundum (RF), and their closest match in GenBank database
| 克隆 Clone | 序列号 GenBank accession no. | BLAST最接近的比对 Closest BLAST match | 重叠区 Overlap (bp) | 相似度 Identity (%) | 克隆数目 Clone No. | ||
|---|---|---|---|---|---|---|---|
| RA | RF | ||||||
| 子囊菌纲 Ascomycota | |||||||
| 061229 | GU256941 | Alternaria alternate (DQ023279) | 608 | 100 | 1 | 3 | |
| 0612271 | GU256942 | — | — | — | 1 | 0 | |
| 061266 | GU256943 | Uncultured Geoglossaceae clone Y43 (DQ273321) | 565 | 90.2 | 8 | 6 | |
| 061626 | GU256944 | Uncultured Rhizoscyphus ericae complex clone K10-16 (AY394687) | 591 | 93.3 | 1 | 5 | |
| 061206 | GU256945 | Uncultured Rhizoscyphus ericae complex clone K10-16 (AY394687) | 550 | 92.5 | 7 | 12 | |
| 061619 | GU256946 | Rhizoscyphus ericae aggregate (AM084704) | 573 | 94.5 | 0 | 2 | |
| 061239 | GU256947 | Rhizoscyphus ericae aggregate (AY394684) | 550 | 87.1 | 3 | 0 | |
| 061286 | GU256948 | Oidiodendron maius strain UAMH 8921 (AF062800) | 527 | 99.1 | 9 | 0 | |
| 061210 | GU256949 | Cryptosporiopsis ericae voucher UAMH 10419 (AY853167) | 599 | 88.1 | 1 | 0 | |
| 061235 | GU256950 | Phialea strobilina strain CBS 643.85 (EF596821) | 571 | 88.3 | 3 | 0 | |
| 061214 | GU256951 | Salal root associated fungus UBCtra1011.13 (AF300726) | 367 | 99.2 | 1 | 0 | |
| 061655 | GU256952 | Mycorrhizal ascomycete of Rhododendron type 3 (AB089663) | 522 | 93.3 | 0 | 4 | |
| 061223 | GU256953 | Mycorrhizal ascomycete of Rhododendron type 2 (AB089660) | 521 | 95.4 | 1 | 0 | |
| 061245 | GU256954 | Mycorrhizal ascomycete of Rhododendron type 2 (AB089660) | 530 | 80.8 | 4 | 0 | |
| 061289 | GU256955 | Penicillium griseofulvum (AB369901) | 577 | 99.6 | 12 | 0 | |
| 061642 | GU256956 | Penicillium sp. 5/97-5 (AJ279476) | 615 | 99.1 | 2 | 2 | |
| 061211 | GU256957 | Paecilomyces sp. JCM 12545 (AB217857) | 632 | 100 | 1 | 0 | |
| 担子菌纲 Basidiomyota | |||||||
| 061221 | GU256958 | Erythrobasidium hasegawianum strain CBS 8253 (AF444522) | 617 | 85.5 | 1 | 0 | |
| 061284 | GU256959 | Erythrobasidium hasegawianum strain CBS 8253 (AF444522) | 635 | 81.6 | 1 | 0 | |
| 061230 | GU256960 | Sporobolomyces yunnanensis (AB030353) | 601 | 99.1 | 10 | 0 | |
| 061277 | GU256961 | Uncultured ectomycorrhiza (Basidiomycota) (AY641466) | 701 | 91.1 | 4 | 0 | |
| 061201 | GU256962 | Cryptococcus terricola strain CBS 6435 (AF444377) | 645 | 100 | 2 | 0 | |
| 061241 | GU256963 | Cryptococcus sp. BF73 (AM901689) | 631 | 98.3 | 1 | 0 | |
| 061624 | GU256964 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 542 | 79.4 | 0 | 14 | |
| 061265 | GU256965 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 542 | 82.4 | 2 | 0 | |
| 061647 | GU256966 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 536 | 89.7 | 0 | 3 | |
| 061692 | GU256967 | Uncultured cf. Tricholoma sp. clone d485.8 (AY254876) | 542 | 80.8 | 0 | 1 | |
| 061632 | GU256968 | Uncultured cf. Tricholoma (AY097046) | 670 | 98.5 | 0 | 2 | |
| 061628 | GU256969 | Uncultured ectomycorrhiza (Tricholomataceae) (AM113458) | 645 | 85.4 | 1 | 20 | |
| 061610 | GU256970 | Tricholoma sp. Umpqua (AF377204) | 725 | 82.6 | 0 | 1 | |
| 061207 | GU256971 | Uncultured mycorrhiza (Sebacinaceae) 4078 (AY634132) | 664 | 84.6 | 5 | 0 | |
| 061220 | GU256972 | Uncultured mycorrhiza (Sebacinales) voucher 04A16 (DQ352049) | 494 | 88.5 | 1 | 0 | |
| 0616101 | GU256973 | Uncultured Sebacinales clone R33 (EF030897) | 412 | 93.0 | 0 | 1 | |
| 061203 | GU256974 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 674 | 92.8 | 2 | 20 | |
| 061209 | GU256975 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 674 | 91.6 | 2 | 0 | |
| 061606 | GU256976 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 671 | 91.6 | 0 | 2 | |
| 061262 | GU256977 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 673 | 90.6 | 4 | 0 | |
| 061607 | GU256978 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 689 | 96.3 | 0 | 1 | |
| 061267 | GU256979 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 679 | 92.1 | 1 | 0 | |
| 061668 | GU256980 | Uncultured Sebacinales clone Rhoner_6300 (EF127237) | 669 | 91.2 | 0 | 2 | |
| 061675 | GU256981 | Gymnopus austrosemihirtipes AWW65 (AY263422) | 796 | 95.3 | 0 | 4 | |
| 真菌 Fungus | 种类数目 No. of ITS taxa | 相对频率 Relative frequency1 | |
|---|---|---|---|
| RA | RF | ||
| Ascomycota | |||
| Helotiales | 10 | 28.2 | 27.6 |
| Eurotiales | 3 | 16.3 | 1.9 |
| Oidiodendron | 1 | 9.9 | 0 |
| Other Ascomycetes | 3 | 5.4 | 2.8 |
| Subtotal | 17 | 59.8 | 32.3 |
| Basidiomycota | |||
| Sebacinales | 10 | 16.3 | 24.8 |
| Erythrobasidiales | 3 | 13.0 | 0 |
| Agaricales | 9 | 7.6 | 42.9 |
| Filobasidiales | 2 | 3.3 | 0 |
| Subtotal | 24 | 40.2 | 67.7 |
Table 2 Relative frequency of fungi orders from root systems of R. argyrophyllum (RA) and R. floribundum(RF)
| 真菌 Fungus | 种类数目 No. of ITS taxa | 相对频率 Relative frequency1 | |
|---|---|---|---|
| RA | RF | ||
| Ascomycota | |||
| Helotiales | 10 | 28.2 | 27.6 |
| Eurotiales | 3 | 16.3 | 1.9 |
| Oidiodendron | 1 | 9.9 | 0 |
| Other Ascomycetes | 3 | 5.4 | 2.8 |
| Subtotal | 17 | 59.8 | 32.3 |
| Basidiomycota | |||
| Sebacinales | 10 | 16.3 | 24.8 |
| Erythrobasidiales | 3 | 13.0 | 0 |
| Agaricales | 9 | 7.6 | 42.9 |
| Filobasidiales | 2 | 3.3 | 0 |
| Subtotal | 24 | 40.2 | 67.7 |
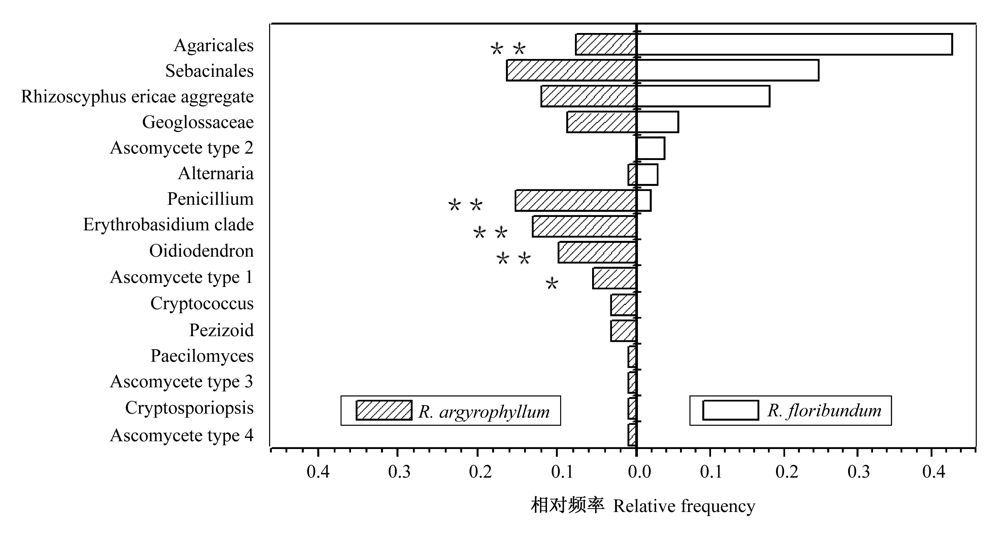
Fig. 1 Relative frequency was calculated as the number of clones from fungi genus, family or order divided by the total number of clones from R. argyrophyllum and R. floribundum. Bars with*were statistically significant (P <0.05); Bars with ** were extremely statistically significant (P<0.01). Ascomycete type 1 Closest BLAST match is AB089660; Ascomycete type 2 Closest BLAST match is AB089663; Ascomycete type 3 Closest BLAST match is AF300726; Ascomycete type 4 is unknown environmental fungi.
| [1] | Allen TR, Millar T, Berch SM, Berbee ML (2003) Culturing and direct DNA extraction find different fungi from the same ericoid mycorrhizal roots. New Phytologist, 160, 255-272. |
| [2] | Berch SM, Allen TR, Berbee ML (2002) Molecular detection, community structure and phylogeny of ericoid mycorrhizal fungi. Plant and Soil, 244, 55-66. |
| [3] |
Bougoure DS, Cairney JWG (2005) Fungi associated with hair roots of Rhododendron lochiae (Ericaceae) in an Australian tropical cloud forest revealed by culturing and culture-independent molecular methods. Environmental Microbiology, 7, 1743-1754.
URL PMID |
| [4] |
Bougoure DS, Parkin PI, Cairney JWG, Alexander IJ, Anderson IC (2007) Diversity of fungi in hair roots of Ericaceae varies along a vegetation gradient. Molecular Ecology, 16, 4624-4636.
DOI URL PMID |
| [5] | Cairney JWG, Ashford AE (2002) Biology of mycorrhizal associations of epacrids (Ericaceae). New Phytologist, 154, 305-326. |
| [6] | Cairney JWG, Meharg AA (2003) Ericoid mycorrhiza: a partnership that exploits harsh edaphic conditions. European Journal of Soil Science, 54, 735-740. |
| [7] | Clarkson DB, Fan Y, Joe H (1993) A remark on algorithm 643: FEXACT: an algorithm for performing fisher's exact test in r × c contingency tables. ACM Transactions on Mathematical Software, 19, 484-488. |
| [8] | Egger KN, Sigler L (1993) Relatedness of the ericoid endophytes Scytalidium vaccinii and Hymenoscyphus ericae inferred from analysis of ribosomal DNA. Mycologia, 85, 219-230. |
| [9] |
Grades M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113-118.
DOI URL PMID |
| [10] | Guo LD, Hyde KD, Liew ECY (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist, 147, 617-630. |
| [11] | Hambleton S, Currah RS (1997) Fungal endophytes from the roots of alpine and boreal Ericaceae. Canadian Journal of Botany, 75, 1570-1581. |
| [12] | Johansson M (2001) Fungal associations of Danish Calluna vulgaris roots with special reference to ericoid mycorrhiza. Plant and Soil, 231, 225-232. |
| [13] | Lereau MJ (1985) Rooting and establishment of in vitro blueberry plants in the presence of mycorrhizal fungi. Acta Horticulturae, 165, 197-201. |
| [14] | Li XL (李雪玲) (2004) A study on the variety of the endo-epiphyte and rhizo-epiphyte of Rhdododendron microphyton. Journal of Chuxiong Normal University (楚雄师范学院学报), 19, 87-91. (in Chinese with English abstract) |
| [15] | Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiology, 5, 28. |
| [16] | Monreal M, Berch SM, Berbee M (1999) Molecular diversity of ericoid mycorrhizal fungi. Canadian Journal of Botany, 77, 1580-1594. |
| [17] | Pearson V, Read DJ (1973) The biology of mycorrhzia in the Ericaceae. II. New Phytologist, 72, 1325-1331. |
| [18] |
Perotto S, ActisPerino E, Perugini J, Bonfante P (1996) Molecular diversity of fungi from ericoid mycorrhizal roots. Molecular Ecology, 5, 123-131.
DOI URL |
| [19] | Perotto S, Peretto R, Faccio A, Schubert A, Varma A, Bonfante P (1995) Ericoid mycorrhizal fungi―cellular and molecular-bases of their interactions with the host-plant. Canadian Journal of Botany, 73, S557-S568. |
| [20] | Read DJ (1996) The structure and function of the ericoid mycorrhizal root. Annals of Botany 77, 365-374. |
| [21] | Renker C, Weibhuhn K, Kellner H, Buscot F (2006) Rationalizing molecular analysis of field-collected roots for assessing diversity of arbuscular mycorrhizal fungi: to pool, or not to pool, that is the question. Mycorrhiza, 16, 525-531. |
| [22] | Selosse MA, Setaro S, Glatard F, Richard F, Urcelay C, Weiss M (2007) Sebacinales are common mycorrhizal associates of Ericaceae. New Phytologist, 174, 864-878. |
| [23] | Setaro S, Weiss M, Oberwinkler F, Kottke I (2006) Sebacinales form ectendomycorrhizas with Cavendishia nobilis, a member of the Andean clade of Ericaceae, in the mountain rain forest of southern Ecuador. New Phytologist, 169, 355-365. |
| [24] | Sigler L, Allan T, Lim SR, Berch S, Berbee M (2005) Two new Cryptosporiopsis species from roots of ericaceous hosts in western North America. Studies in Mycology, 53, 53-62. |
| [25] | Smith SE, Read DJ (1997) Mycorrhizal Symbiosis. Academic Press, London. |
| [26] | Straker CJ (1996) Ericoid mycorrhiza: ecological and host specificity. Mycorrhiza, 6, 215-225. |
| [27] | Usuki F, Abe JP, Kakishima M (2003) Diversity of ericoid mycorrhizal fungi isolated from hair roots of Rhododendron obtusum var. kaempferi in a Japanese red pine forest. Mycoscience, 44, 97-102. |
| [28] | Vralstad T, Myhre E, Schumacher T (2002) Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytologist, 155, 131-148. |
| [29] |
Weiss M, Selosse MA, Rexer KH, Urban A, Oberwinkler F (2004) Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycological Research, 108, 1003-1010.
URL PMID |
| [30] | Wu CH (吴重华), Wang JR (王吉忍), Yang JX (杨俊秀), Hu CD (胡崇德) (2000) A study on Rhododendron clementiae Forrest ex W.W. Smith Subsp. aureodorsale W. P. Fang’Mycorrhiaze in the Nature Preserve of the Taibai Mountains. Journal of Northwest Forestry University (西北林学院学报), 15(3), 68-70. (in Chinese with English abstract) |
| [31] | Zhang C, Yin L, Dai S (2009) Diversity of root-associated fungal endophytes in Rhododendron fortunei in subtropical forests of China. Mycorrhiza, 19, 417-423. |
| [32] | Zhang YH, Zhuang WY (2004) Phylogenetic relationships of some members in the genus Hymenoscyphus (Ascomycetes, Helotiales). Nova Hedwigia, 78, 475-484. |
| [33] | Zijlstra JD, Van't Hof P, Baar J, Verkley GJM, Summerbell RC, Paradi I, Braakhekke WG, Berendse F (2005) Diversity of symbiotic root endophytes of the Helotiales in ericaceous plants and the grass, Deschampsia flexuosa. Studies in Mycology, 53, 147-162. |
| [1] | Min Luo, Yongchuan Yang, Cheng Jin, Lihua Zhou, Yuxiao Long. Composition profile and response to human activity of mammals in the urban forests of central Chongqing [J]. Biodiv Sci, 2025, 33(5): 24402-. |
| [2] | Zhang Songqi, Lu Yi, Chen Bingyao, Yang Guang, Wang Yanping, Chen Chuanwu. A dataset on the morphological, life-history, and ecological traits of cetaceans worldwide [J]. Biodiv Sci, 2025, 33(2): 24442-. |
| [3] | Zhao Yifan, Wang Yanping. A database of life-history, ecological, and biogeographical traits of snakes worldwide [J]. Biodiv Sci, 2025, 33(2): 24476-. |
| [4] | Chuan Jin, Zijia Zhang, Kai Di, Weirong Zhang, Dong Qiao, Siyuan Cheng, Zhongmin Hu. A dataset on fluorescence, photosynthesis gas exchange, and leaf traits of Hainan tropical rainforest plant species [J]. Biodiv Sci, 2024, 32(9): 24139-. |
| [5] | Jing Chen, Bingchang Zhang, Yanjin Liu, Jie Wu, Kang Zhao, Jiao Ming. Diversity of Leptolyngbya-like cyanobacteria in biocrusts in desert area [J]. Biodiv Sci, 2024, 32(9): 24186-. |
| [6] | Fei Duan, Mingzhang Liu, Hongliang Bu, Le Yu, Sheng Li. Effects of urbanization on bird community composition and functional traits: A case study of the Beijing-Tianjin-Hebei region [J]. Biodiv Sci, 2024, 32(8): 23473-. |
| [7] | Hongyu Niu, Lu Chen, Hengyue Zhao, Gulzar Abdukirim, Hongmao Zhang. Effects of urbanization on animals: From community to individual level [J]. Biodiv Sci, 2024, 32(8): 23489-. |
| [8] | Yujin Cui, Wanying Li, Qingqing Zhou, Heng Zhao, Fang Wu, Yuan Yuan. Diversity and community composition of epiphytic fungi in the phyllosphere of Pinus tabuliformis and Euonymus japonicus in Beijing, northern China [J]. Biodiv Sci, 2024, 32(7): 23498-. |
| [9] | Bohan Zheng, Xinyao Chen, Jian Ni. A dataset on the plant growth form and life form of vascular plants in China [J]. Biodiv Sci, 2024, 32(7): 23468-. |
| [10] | Weijie Shu, Hua He, Luo Zeng, Zhirong Gu, Dunyan Tan, Xiaochen Yang. Spatial distribution and sexual dimorphism of dioecious Arisaema erubescens [J]. Biodiv Sci, 2024, 32(6): 24084-. |
| [11] | Yunyue Peng, Tong Jin, Xiaoquan Zhang. Biodiversity credits: Concepts, principles, transactions and challenges [J]. Biodiv Sci, 2024, 32(2): 23300-. |
| [12] | Jialong Ren, Yongzhen Wang, Yilin Feng, Wenzhi Zhao, Qihan Yan, Chang Qin, Jing Fang, Weidong Xin, Jiliang Liu. Beetle data set collected by pitfall trapping in the gobi desert of the Hexi Corridor [J]. Biodiv Sci, 2024, 32(2): 23375-. |
| [13] | Jiayi Feng, Juyu Lian, Yujun Feng, Dongxu Zhang, Honglin Cao, Wanhui Ye. Effects of vertical stratification on community structure and functions in a subtropical, evergreen broad-leaved forest in the Dinghushan National Nature Reserve [J]. Biodiv Sci, 2024, 32(12): 24306-. |
| [14] | Jiang Wang, Yifan Zhao, Yanfu Qu, Caiwen Zhang, Liang Zhang, Chuanwu Chen, Yanping Wang. A dataset of the morphological, life-history, and ecological traits of snakes in China [J]. Biodiv Sci, 2023, 31(7): 23126-. |
| [15] | Renxiu Yao, Yan Chen, Xiaoqin Lü, Jianghu Wang, Fujun Yang, Xiaoyue Wang. Altitude-related environmental factors shape the phenotypic characteristics and chemical profile of Rhododendron [J]. Biodiv Sci, 2023, 31(2): 22259-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()