



Biodiv Sci ›› 2021, Vol. 29 ›› Issue (12): 1638-1649. DOI: 10.17520/biods.2021249 cstr: 32101.14.biods.2021249
Special Issue: 物种形成与系统进化
• Original Papers: Plant Diversity • Previous Articles Next Articles
Xiangxiang Chen1,2, Zhongshuai Gai1,2, Juntuan Zhai1,2,3, Jindong Xu4, Peipei Jiao1,2,3, Zhihua Wu4,*( ), Zhijun Li1,2,3,*(
), Zhijun Li1,2,3,*( )
)
Received:2021-06-23
Accepted:2021-09-30
Online:2021-12-20
Published:2021-11-05
Contact:
Zhihua Wu,Zhijun Li
About author:First author contact:#Co-first authors
Xiangxiang Chen, Zhongshuai Gai, Juntuan Zhai, Jindong Xu, Peipei Jiao, Zhihua Wu, Zhijun Li. Genetic diversity and construction of core conservation units of the natural populations of Populus euphratica in Northwest China[J]. Biodiv Sci, 2021, 29(12): 1638-1649.
| 地区 Region | 样点 Site | 群体 Population | 经纬度 Location | 海拔 Altitude (m) | ||
|---|---|---|---|---|---|---|
| 南疆阿克苏 Aksu, SX | 柯坪县 Kalpin County | AKS-KPX | 79.52° E, 40.22° N | 1,042-1,095 | ||
| 阿瓦提县 Awat County | AKS-AWT | 80.94° E, 40.36° N | 1,000-1,036 | |||
| 温宿县 Wensu County | AKS-WSX | 80.79° E, 41.21° N | 1,104-1,140 | |||
| 沙雅县 Shaya County | AKS-SYX | 83.42° E, 41.59° N | 929-989 | |||
| 库车县 Kuqa County | AKS-KCS | 83.86° E, 41.28° N | 917-934 | |||
| 拜城县 Baicheng County | AKS-BCX | 81.81° E, 41.66° N | 1,231-1,256 | |||
| 阿拉尔市 Alaer City | AKS-ALE | 81.03° E, 40.51° N | 1,012-1,038 | |||
| 南疆喀什 Kashgar, SX | 巴楚县 Marabishi County | KS-BCX | 78.31° E, 39.77° N | 1,121-1,137 | ||
| 麦盖提县 Makit County | KS-MGTX | 78.24° E, 39.36° N | 1,132-1,156 | |||
| 岳普湖县 Yopurga County | KS-YPHX | 77.24° E, 39.18° N | 1,166-1,246 | |||
| 伽师县 Payzawat County | KS-JSX | 77.38° E, 39.65° N | 1,161-1,174 | |||
| 疏勒县 Shule County | KS-SLX | 76.18° E, 30.17° N | 1,259-1,279 | |||
| 莎车县 Yarkant County | KS-SCX | 77.36° E, 38.40° N | 1,187-1,220 | |||
| 叶城县 Yecheng County | KS-YCX | 77.58° E, 38.18° N | 1,230-1,299 | |||
| 南疆和田 Hotan, SX | 皮山县 Pishan County | HT-PSX | 79.01° E, 37.62° N | 1,290-1,328 | ||
| 墨玉县 Karakax County | HT-MYX | 90.19° E, 37.77° N | 1,239-1,290 | |||
| 洛浦县 Lop County | HT-LPX | 80.12° E, 37.43° N | 1,101-1,284 | |||
| 策勒县 Qira County | HT-CLX | 81.03° E, 36.94° N | 1,394-1,457 | |||
| 于田县 Yutian County | HT-YTX | 81.41° E, 37.60° N | 1,263-1,452 | |||
| 民丰县 Minfeng County | HT-MFX | 82.79° E, 37.73° N | 1,278-1,387 | |||
| 南疆巴音郭楞蒙古自治州 Bayingol Mongolian Autonomous Prefecture, SX | 且末县 Qiemo County | BZ-QMX | 84.09° E, 37.73° N | 1,088-1,315 | ||
| 若羌县 Ruoqiang County | BaZ-RQX | 87.12° E, 38.78° N | 801-984 | |||
| 尉犁县 Yuli County | BaZ-YLX | 85.77° E, 41.14° N | 841-931 | |||
| 库尔勒市 Korla City | BaZ-KEL | 85.78° E, 41.53° N | 884-895 | |||
| 和硕县 Heshuo City | BaZ-HSX | 86.85° E, 42.17° N | 1,047-1,084 | |||
| 轮台县 Bugur County | BaZ-LTX | 84.25° E, 41.26° N | 905-969 | |||
| 甘肃 Gansu | 民勤县 Minqin County | GS-MQX | 103.1° E, 38.65° N | 1,283-1,323 | ||
| 金塔县 Jinta County | GS-JTX | 98.87° E, 39.93° N | 1,191-1,284 | |||
| 瓜州县 Guazhou County | GS-GZX | 96.05° E, 40.54° N | 1,027-1,218 | |||
| 敦煌县 Dunhuang County | GS-DHX | 94.83° E, 40.17° N | 1,033-1,140 | |||
| 阿克塞哈萨克族自治县 Kazak Autonomous County of Aksay | GS-AKS | 93.43° E, 39.78° N | 1,447-1,493 | |||
| 宁夏 Ningxia | 中卫市 Zhongwei City | NIX-ZW | 105.12° E, 37.59° N | 1,207-1,220 | ||
| 内蒙古 Inner Mongolia | 额济纳旗 Ejina | NMG-EJN | 101.08° E, 41.97° N | 813-926 | ||
| 四子王旗 Siziwang Banner | NMG-SZWQ | 111.27° E, 42.83° N | 901-963 | |||
| 青海 Qinghai | 格尔木市 Geermu City | QH-GEM | 94.33° E, 36.44° N | 2,709-2,936 | ||
| 北疆吐鲁番 Turpan, NX | 托克逊县 Tuokexun County | TLF-TKX | 88.17° E, 43.21° N | 1,128-1,152 | ||
| 北疆哈密 Hami, NX | 伊州区 Yizhou District | HM-YZQ | 93.08° E, 42.59° N | 390-405 | ||
| 伊吾县 Yiwu County | HM-YWX | 95.25° E, 43.66° N | 456-489 | |||
| 巴里坤县 Barkol County | HM-BLKX | 93.97° E, 43.97° N | 1,049-1,120 | |||
| 北疆昌吉 Changji, NX | 木垒哈萨克自治县 Mulei Kazak Autonomous County | CJ-MLX | 91.29° E, 44.82° N | 730-756 | ||
| 吉木萨尔县 Jimusaer County | CJ-JMSE | 89.01° E, 44.07° N | 670-676 | |||
| 呼图壁县 Hutubi County | CJ-HTBX | 86.90° E, 44.52° N | 359-401 | |||
| 玛纳斯县 Manasi County | CJ-MNSX | 86.87° E, 44.66° N | 353-362 | |||
| 北疆阿勒泰 Altay, NX | 哈巴河县 Habahe County | ALT-HBH | 86.26° E, 47.92° N | 431-456 | ||
| 福海县 Fuhai County | ALT-FHX | 87.78° E, 47.21° N | 489-494 | |||
| 布尔津县 Burqin County | ALT-BEJ | 86.94° E, 47.66° N | 466-494 | |||
| 富蕴县 Fuyun County | ALT-FYX | 88.75° E, 46.91° N | 677-724 | |||
| 北疆博州 Bortala Mongol Autonomous Prefecture, NX | 阿拉山口市 Alashankou City | BoZ-ALSK | 82.97° E, 45.01° N | 194-206 | ||
| 精河县 Jinghe County | BoZ-JHX | 83.43° E, 44.91° N | 203-217 | |||
| 北疆塔城 Tarbagatay, NX | 沙湾县 Shawan County | TC-SWX | 85.77° E, 44.88° N | 312-328 | ||
| 乌苏市 Wusu City | TC-WUSU | 83.83° E, 44.80° N | 260-268 | |||
| 和丰县 Hefeng County | TC-HFX | 85.27° E, 46.26° N | 469-477 | |||
| 托里县 Toli County | TC-TLX | 85.32° E, 46.15° N | 426-431 | |||
| 北疆伊犁哈萨克自治州 Ili Kazak Autonomous Prefecture, NX | 霍城县 Huocheng County | YL-HCX | 80.55° E, 44.09° N | 631-638 | ||
| 察布查尔锡伯自治县 Qapqal Xibe Autonomous County | YL-CX | 80.69° E, 43.84° N | 528-542 | |||
| 北疆克拉玛依 Karamay, NX | 克拉玛依区 Karamay District | KLMYS | 85.14° E, 45.33° N | 271-278 | ||
| 乌尔禾区 Urho District | KLMY-WEH | 85.67° E, 46.13° N | 296-302 | |||
| 北疆乌鲁木齐 Urumqi, NX | 乌鲁木齐市 Urumqi City | WLMQ | 88.35° E, 43.28° N | 936-969 | ||
Table 1 Sampling information of 58 Populus euphratica populations in this study
| 地区 Region | 样点 Site | 群体 Population | 经纬度 Location | 海拔 Altitude (m) | ||
|---|---|---|---|---|---|---|
| 南疆阿克苏 Aksu, SX | 柯坪县 Kalpin County | AKS-KPX | 79.52° E, 40.22° N | 1,042-1,095 | ||
| 阿瓦提县 Awat County | AKS-AWT | 80.94° E, 40.36° N | 1,000-1,036 | |||
| 温宿县 Wensu County | AKS-WSX | 80.79° E, 41.21° N | 1,104-1,140 | |||
| 沙雅县 Shaya County | AKS-SYX | 83.42° E, 41.59° N | 929-989 | |||
| 库车县 Kuqa County | AKS-KCS | 83.86° E, 41.28° N | 917-934 | |||
| 拜城县 Baicheng County | AKS-BCX | 81.81° E, 41.66° N | 1,231-1,256 | |||
| 阿拉尔市 Alaer City | AKS-ALE | 81.03° E, 40.51° N | 1,012-1,038 | |||
| 南疆喀什 Kashgar, SX | 巴楚县 Marabishi County | KS-BCX | 78.31° E, 39.77° N | 1,121-1,137 | ||
| 麦盖提县 Makit County | KS-MGTX | 78.24° E, 39.36° N | 1,132-1,156 | |||
| 岳普湖县 Yopurga County | KS-YPHX | 77.24° E, 39.18° N | 1,166-1,246 | |||
| 伽师县 Payzawat County | KS-JSX | 77.38° E, 39.65° N | 1,161-1,174 | |||
| 疏勒县 Shule County | KS-SLX | 76.18° E, 30.17° N | 1,259-1,279 | |||
| 莎车县 Yarkant County | KS-SCX | 77.36° E, 38.40° N | 1,187-1,220 | |||
| 叶城县 Yecheng County | KS-YCX | 77.58° E, 38.18° N | 1,230-1,299 | |||
| 南疆和田 Hotan, SX | 皮山县 Pishan County | HT-PSX | 79.01° E, 37.62° N | 1,290-1,328 | ||
| 墨玉县 Karakax County | HT-MYX | 90.19° E, 37.77° N | 1,239-1,290 | |||
| 洛浦县 Lop County | HT-LPX | 80.12° E, 37.43° N | 1,101-1,284 | |||
| 策勒县 Qira County | HT-CLX | 81.03° E, 36.94° N | 1,394-1,457 | |||
| 于田县 Yutian County | HT-YTX | 81.41° E, 37.60° N | 1,263-1,452 | |||
| 民丰县 Minfeng County | HT-MFX | 82.79° E, 37.73° N | 1,278-1,387 | |||
| 南疆巴音郭楞蒙古自治州 Bayingol Mongolian Autonomous Prefecture, SX | 且末县 Qiemo County | BZ-QMX | 84.09° E, 37.73° N | 1,088-1,315 | ||
| 若羌县 Ruoqiang County | BaZ-RQX | 87.12° E, 38.78° N | 801-984 | |||
| 尉犁县 Yuli County | BaZ-YLX | 85.77° E, 41.14° N | 841-931 | |||
| 库尔勒市 Korla City | BaZ-KEL | 85.78° E, 41.53° N | 884-895 | |||
| 和硕县 Heshuo City | BaZ-HSX | 86.85° E, 42.17° N | 1,047-1,084 | |||
| 轮台县 Bugur County | BaZ-LTX | 84.25° E, 41.26° N | 905-969 | |||
| 甘肃 Gansu | 民勤县 Minqin County | GS-MQX | 103.1° E, 38.65° N | 1,283-1,323 | ||
| 金塔县 Jinta County | GS-JTX | 98.87° E, 39.93° N | 1,191-1,284 | |||
| 瓜州县 Guazhou County | GS-GZX | 96.05° E, 40.54° N | 1,027-1,218 | |||
| 敦煌县 Dunhuang County | GS-DHX | 94.83° E, 40.17° N | 1,033-1,140 | |||
| 阿克塞哈萨克族自治县 Kazak Autonomous County of Aksay | GS-AKS | 93.43° E, 39.78° N | 1,447-1,493 | |||
| 宁夏 Ningxia | 中卫市 Zhongwei City | NIX-ZW | 105.12° E, 37.59° N | 1,207-1,220 | ||
| 内蒙古 Inner Mongolia | 额济纳旗 Ejina | NMG-EJN | 101.08° E, 41.97° N | 813-926 | ||
| 四子王旗 Siziwang Banner | NMG-SZWQ | 111.27° E, 42.83° N | 901-963 | |||
| 青海 Qinghai | 格尔木市 Geermu City | QH-GEM | 94.33° E, 36.44° N | 2,709-2,936 | ||
| 北疆吐鲁番 Turpan, NX | 托克逊县 Tuokexun County | TLF-TKX | 88.17° E, 43.21° N | 1,128-1,152 | ||
| 北疆哈密 Hami, NX | 伊州区 Yizhou District | HM-YZQ | 93.08° E, 42.59° N | 390-405 | ||
| 伊吾县 Yiwu County | HM-YWX | 95.25° E, 43.66° N | 456-489 | |||
| 巴里坤县 Barkol County | HM-BLKX | 93.97° E, 43.97° N | 1,049-1,120 | |||
| 北疆昌吉 Changji, NX | 木垒哈萨克自治县 Mulei Kazak Autonomous County | CJ-MLX | 91.29° E, 44.82° N | 730-756 | ||
| 吉木萨尔县 Jimusaer County | CJ-JMSE | 89.01° E, 44.07° N | 670-676 | |||
| 呼图壁县 Hutubi County | CJ-HTBX | 86.90° E, 44.52° N | 359-401 | |||
| 玛纳斯县 Manasi County | CJ-MNSX | 86.87° E, 44.66° N | 353-362 | |||
| 北疆阿勒泰 Altay, NX | 哈巴河县 Habahe County | ALT-HBH | 86.26° E, 47.92° N | 431-456 | ||
| 福海县 Fuhai County | ALT-FHX | 87.78° E, 47.21° N | 489-494 | |||
| 布尔津县 Burqin County | ALT-BEJ | 86.94° E, 47.66° N | 466-494 | |||
| 富蕴县 Fuyun County | ALT-FYX | 88.75° E, 46.91° N | 677-724 | |||
| 北疆博州 Bortala Mongol Autonomous Prefecture, NX | 阿拉山口市 Alashankou City | BoZ-ALSK | 82.97° E, 45.01° N | 194-206 | ||
| 精河县 Jinghe County | BoZ-JHX | 83.43° E, 44.91° N | 203-217 | |||
| 北疆塔城 Tarbagatay, NX | 沙湾县 Shawan County | TC-SWX | 85.77° E, 44.88° N | 312-328 | ||
| 乌苏市 Wusu City | TC-WUSU | 83.83° E, 44.80° N | 260-268 | |||
| 和丰县 Hefeng County | TC-HFX | 85.27° E, 46.26° N | 469-477 | |||
| 托里县 Toli County | TC-TLX | 85.32° E, 46.15° N | 426-431 | |||
| 北疆伊犁哈萨克自治州 Ili Kazak Autonomous Prefecture, NX | 霍城县 Huocheng County | YL-HCX | 80.55° E, 44.09° N | 631-638 | ||
| 察布查尔锡伯自治县 Qapqal Xibe Autonomous County | YL-CX | 80.69° E, 43.84° N | 528-542 | |||
| 北疆克拉玛依 Karamay, NX | 克拉玛依区 Karamay District | KLMYS | 85.14° E, 45.33° N | 271-278 | ||
| 乌尔禾区 Urho District | KLMY-WEH | 85.67° E, 46.13° N | 296-302 | |||
| 北疆乌鲁木齐 Urumqi, NX | 乌鲁木齐市 Urumqi City | WLMQ | 88.35° E, 43.28° N | 936-969 | ||
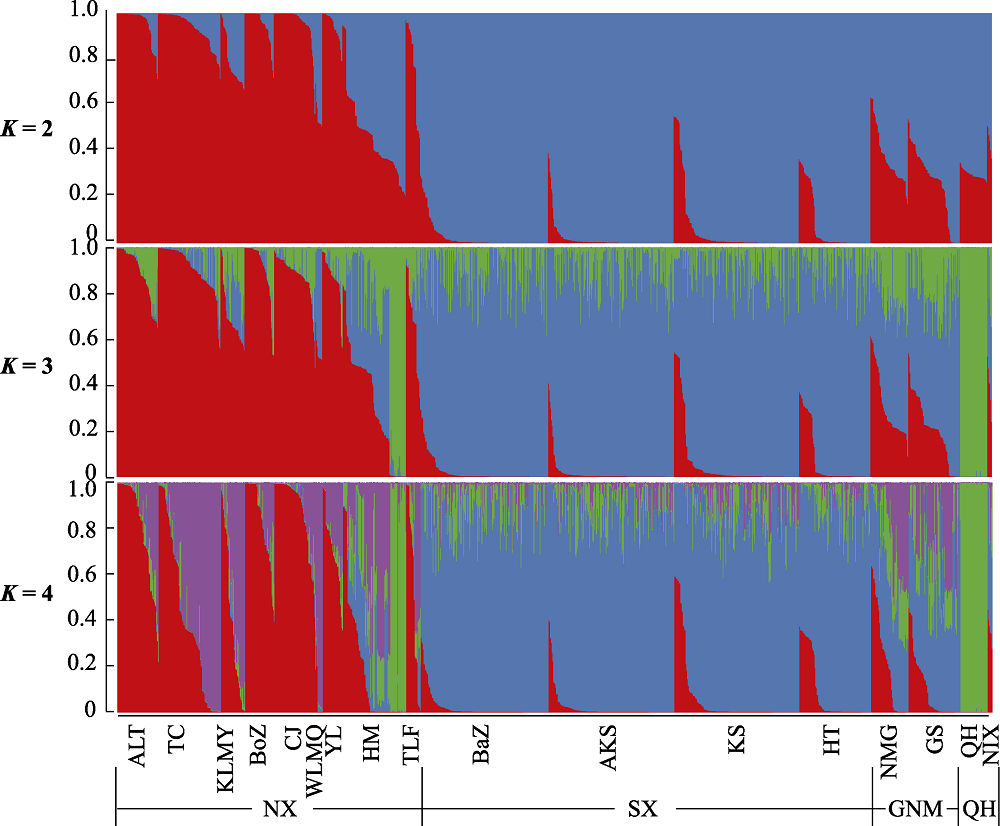
Fig. 1 The structural analysis of Populus euphratica populations in China. SX, Southern Xinjiang; NX, Northern Xinjiang; QH, Qinghai; GNM, Gansu, Ningxia, and Inner Mongolia. Population codes see Table 1.
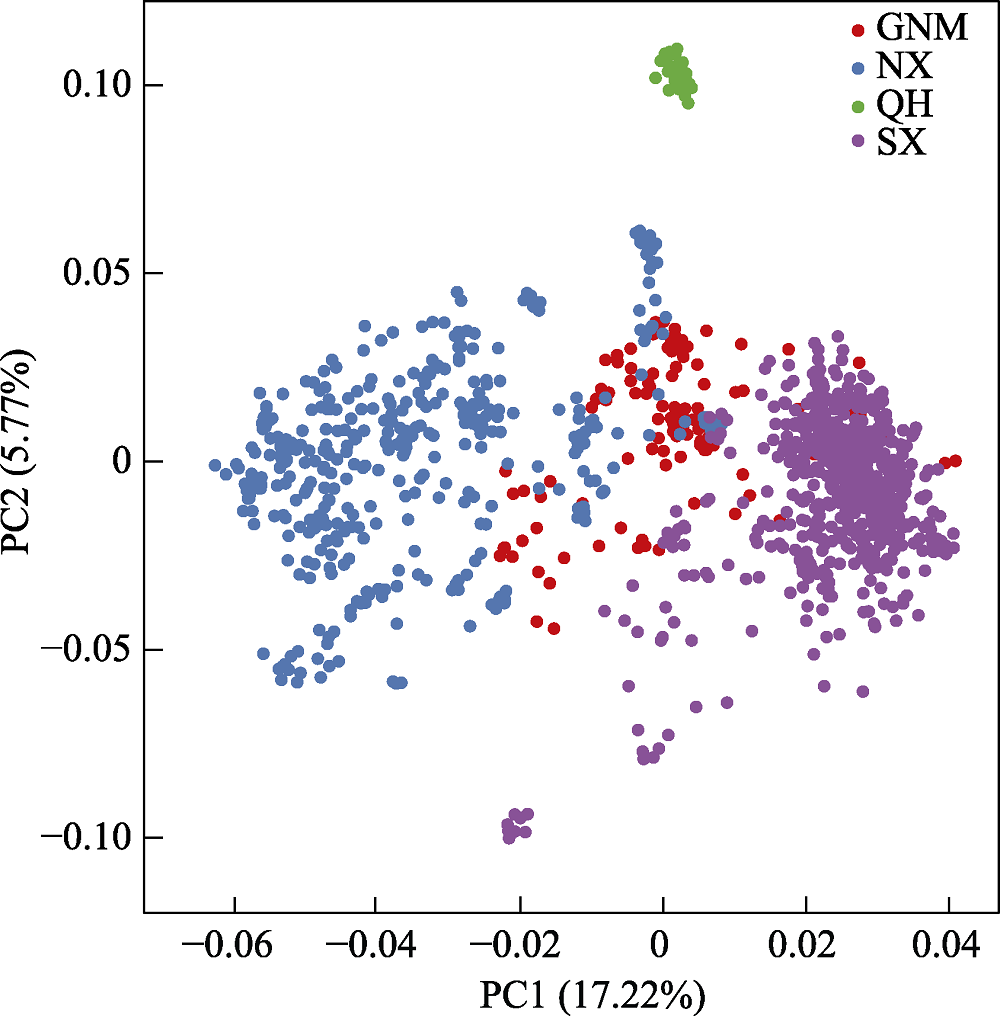
Fig. 2 The PCA analysis of Populus euphratica populations in China. SX, Southern Xinjiang; NX, Northern Xinjiang; QH, Qinghai; GNM, Gansu, Ningxia, and Inner Mongolia.
| 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC | 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC |
|---|---|---|---|---|---|---|---|---|---|
| KS-MGTX | 0.294 | 0.262 | 0.443 | 0.235 | BoZ-JHX | 0.217 | 0.217 | 0.327 | 0.174 |
| AKS-ALE | 0.278 | 0.270 | 0.420 | 0.223 | TLF-TKX | 0.213 | 0.230 | 0.321 | 0.171 |
| KS-YPHX | 0.266 | 0.261 | 0.406 | 0.215 | AKS-WSX | 0.211 | 0.238 | 0.319 | 0.169 |
| HT-LPX | 0.261 | 0.264 | 0.390 | 0.208 | GS-DHX | 0.210 | 0.242 | 0.310 | 0.165 |
| HT-PSX | 0.248 | 0.255 | 0.378 | 0.200 | KLMYS | 0.210 | 0.211 | 0.315 | 0.167 |
| HT-YTX | 0.246 | 0.244 | 0.373 | 0.198 | TC-WUSU | 0.208 | 0.239 | 0.314 | 0.167 |
| KS-JSX | 0.246 | 0.243 | 0.378 | 0.199 | ALT-BEJ | 0.205 | 0.219 | 0.308 | 0.164 |
| NMG-EJN | 0.244 | 0.238 | 0.368 | 0.196 | TC-SWX | 0.204 | 0.229 | 0.306 | 0.164 |
| KS-BCX | 0.244 | 0.245 | 0.374 | 0.197 | HT-CLX | 0.203 | 0.258 | 0.304 | 0.161 |
| HT-MFX | 0.241 | 0.259 | 0.368 | 0.195 | GS-AKS | 0.200 | 0.249 | 0.303 | 0.161 |
| KS-SLX | 0.240 | 0.248 | 0.365 | 0.193 | CJ-HTB | 0.198 | 0.255 | 0.297 | 0.159 |
| NX-ZW | 0.236 | 0.261 | 0.351 | 0.188 | YL-HCX | 0.184 | 0.239 | 0.274 | 0.147 |
| AKS-SYX | 0.234 | 0.233 | 0.358 | 0.189 | CJ-JMSE | 0.177 | 0.199 | 0.266 | 0.142 |
| BaZ-KEL | 0.234 | 0.253 | 0.355 | 0.188 | YL-CX | 0.177 | 0.222 | 0.271 | 0.145 |
| GS-GZX | 0.231 | 0.245 | 0.347 | 0.185 | HM-BLKX | 0.176 | 0.225 | 0.255 | 0.137 |
| HT-MYX | 0.230 | 0.241 | 0.350 | 0.185 | TC-HFX | 0.174 | 0.221 | 0.261 | 0.139 |
| BaZ-HSX | 0.230 | 0.250 | 0.344 | 0.183 | ALT-FHX | 0.174 | 0.214 | 0.266 | 0.142 |
| KS-SCX | 0.228 | 0.246 | 0.347 | 0.184 | ALT-HBH | 0.174 | 0.210 | 0.267 | 0.142 |
| BaZ-RQX | 0.228 | 0.244 | 0.347 | 0.183 | KLMY-WEH | 0.172 | 0.245 | 0.259 | 0.137 |
| AKS-AWT | 0.228 | 0.237 | 0.351 | 0.185 | BoZ-ALSK | 0.167 | 0.208 | 0.262 | 0.138 |
| GS-JTX | 0.228 | 0.243 | 0.342 | 0.182 | HM-YZQ | 0.164 | 0.239 | 0.247 | 0.131 |
| BaZ-LTX | 0.226 | 0.232 | 0.340 | 0.180 | AKT-FYX | 0.154 | 0.239 | 0.225 | 0.120 |
| AKS-KCX | 0.225 | 0.228 | 0.340 | 0.181 | GS-MQX | 0.145 | 0.274 | 0.205 | 0.110 |
| AKS-BCX | 0.224 | 0.237 | 0.345 | 0.181 | CJ-MLX | 0.142 | 0.253 | 0.204 | 0.109 |
| CJ-MNS | 0.224 | 0.254 | 0.333 | 0.178 | WLMQ | 0.140 | 0.218 | 0.214 | 0.112 |
| BaZ-YLX | 0.222 | 0.245 | 0.339 | 0.179 | NMG-SZWQ | 0.128 | 0.237 | 0.182 | 0.098 |
| KS-YCX | 0.222 | 0.247 | 0.335 | 0.178 | QH-GEM | 0.126 | 0.233 | 0.181 | 0.097 |
| AKS-KPX | 0.221 | 0.243 | 0.333 | 0.177 | TC-TLX | 0.109 | 0.204 | 0.153 | 0.082 |
| BaZ-QMX | 0.218 | 0.241 | 0.329 | 0.175 | 平均 Mean | 0.208 | 0.239 | 0.314 | 0.167 |
| HM-YWX | 0.217 | 0.238 | 0.328 | 0.174 | 标准差 SD | 0.039 | 0.016 | 0.061 | 0.032 |
Table 2 Genetic diversity analysis of 58 natural populations of Populus euphratica. He, Expected heterozygosity; Ho, Observed heterozygosity; I, Shannon’s information index; PIC, Polymorphism information content. Population codes see Table 1.
| 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC | 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC |
|---|---|---|---|---|---|---|---|---|---|
| KS-MGTX | 0.294 | 0.262 | 0.443 | 0.235 | BoZ-JHX | 0.217 | 0.217 | 0.327 | 0.174 |
| AKS-ALE | 0.278 | 0.270 | 0.420 | 0.223 | TLF-TKX | 0.213 | 0.230 | 0.321 | 0.171 |
| KS-YPHX | 0.266 | 0.261 | 0.406 | 0.215 | AKS-WSX | 0.211 | 0.238 | 0.319 | 0.169 |
| HT-LPX | 0.261 | 0.264 | 0.390 | 0.208 | GS-DHX | 0.210 | 0.242 | 0.310 | 0.165 |
| HT-PSX | 0.248 | 0.255 | 0.378 | 0.200 | KLMYS | 0.210 | 0.211 | 0.315 | 0.167 |
| HT-YTX | 0.246 | 0.244 | 0.373 | 0.198 | TC-WUSU | 0.208 | 0.239 | 0.314 | 0.167 |
| KS-JSX | 0.246 | 0.243 | 0.378 | 0.199 | ALT-BEJ | 0.205 | 0.219 | 0.308 | 0.164 |
| NMG-EJN | 0.244 | 0.238 | 0.368 | 0.196 | TC-SWX | 0.204 | 0.229 | 0.306 | 0.164 |
| KS-BCX | 0.244 | 0.245 | 0.374 | 0.197 | HT-CLX | 0.203 | 0.258 | 0.304 | 0.161 |
| HT-MFX | 0.241 | 0.259 | 0.368 | 0.195 | GS-AKS | 0.200 | 0.249 | 0.303 | 0.161 |
| KS-SLX | 0.240 | 0.248 | 0.365 | 0.193 | CJ-HTB | 0.198 | 0.255 | 0.297 | 0.159 |
| NX-ZW | 0.236 | 0.261 | 0.351 | 0.188 | YL-HCX | 0.184 | 0.239 | 0.274 | 0.147 |
| AKS-SYX | 0.234 | 0.233 | 0.358 | 0.189 | CJ-JMSE | 0.177 | 0.199 | 0.266 | 0.142 |
| BaZ-KEL | 0.234 | 0.253 | 0.355 | 0.188 | YL-CX | 0.177 | 0.222 | 0.271 | 0.145 |
| GS-GZX | 0.231 | 0.245 | 0.347 | 0.185 | HM-BLKX | 0.176 | 0.225 | 0.255 | 0.137 |
| HT-MYX | 0.230 | 0.241 | 0.350 | 0.185 | TC-HFX | 0.174 | 0.221 | 0.261 | 0.139 |
| BaZ-HSX | 0.230 | 0.250 | 0.344 | 0.183 | ALT-FHX | 0.174 | 0.214 | 0.266 | 0.142 |
| KS-SCX | 0.228 | 0.246 | 0.347 | 0.184 | ALT-HBH | 0.174 | 0.210 | 0.267 | 0.142 |
| BaZ-RQX | 0.228 | 0.244 | 0.347 | 0.183 | KLMY-WEH | 0.172 | 0.245 | 0.259 | 0.137 |
| AKS-AWT | 0.228 | 0.237 | 0.351 | 0.185 | BoZ-ALSK | 0.167 | 0.208 | 0.262 | 0.138 |
| GS-JTX | 0.228 | 0.243 | 0.342 | 0.182 | HM-YZQ | 0.164 | 0.239 | 0.247 | 0.131 |
| BaZ-LTX | 0.226 | 0.232 | 0.340 | 0.180 | AKT-FYX | 0.154 | 0.239 | 0.225 | 0.120 |
| AKS-KCX | 0.225 | 0.228 | 0.340 | 0.181 | GS-MQX | 0.145 | 0.274 | 0.205 | 0.110 |
| AKS-BCX | 0.224 | 0.237 | 0.345 | 0.181 | CJ-MLX | 0.142 | 0.253 | 0.204 | 0.109 |
| CJ-MNS | 0.224 | 0.254 | 0.333 | 0.178 | WLMQ | 0.140 | 0.218 | 0.214 | 0.112 |
| BaZ-YLX | 0.222 | 0.245 | 0.339 | 0.179 | NMG-SZWQ | 0.128 | 0.237 | 0.182 | 0.098 |
| KS-YCX | 0.222 | 0.247 | 0.335 | 0.178 | QH-GEM | 0.126 | 0.233 | 0.181 | 0.097 |
| AKS-KPX | 0.221 | 0.243 | 0.333 | 0.177 | TC-TLX | 0.109 | 0.204 | 0.153 | 0.082 |
| BaZ-QMX | 0.218 | 0.241 | 0.329 | 0.175 | 平均 Mean | 0.208 | 0.239 | 0.314 | 0.167 |
| HM-YWX | 0.217 | 0.238 | 0.328 | 0.174 | 标准差 SD | 0.039 | 0.016 | 0.061 | 0.032 |
| 来源 Source | 自由度 Degree of freedom (df) | 平方和 Sum of square (SS) | 均方 Mean square (MS) | 方差分量 Variance component | 方差分量占比 Proportion (%) |
|---|---|---|---|---|---|
| 群体间 Among populations | 57 | 93,766.13 | 1,645.02 | 36.40 | 17.88 |
| 个体间 Among individuals | 1,103 | 215,156.92 | 195.07 | 27.96 | 13.74 |
| 个体内 Within individual | 1,161 | 161,558 | 139.15 | 139.15 | 68.38 |
| 总计 Total | 2,321 | 470,481.05 | - | 203.51 | 100 |
Table 3 Analysis of molecular variance of genetic variation within and among populations of Populus euphratica
| 来源 Source | 自由度 Degree of freedom (df) | 平方和 Sum of square (SS) | 均方 Mean square (MS) | 方差分量 Variance component | 方差分量占比 Proportion (%) |
|---|---|---|---|---|---|
| 群体间 Among populations | 57 | 93,766.13 | 1,645.02 | 36.40 | 17.88 |
| 个体间 Among individuals | 1,103 | 215,156.92 | 195.07 | 27.96 | 13.74 |
| 个体内 Within individual | 1,161 | 161,558 | 139.15 | 139.15 | 68.38 |
| 总计 Total | 2,321 | 470,481.05 | - | 203.51 | 100 |
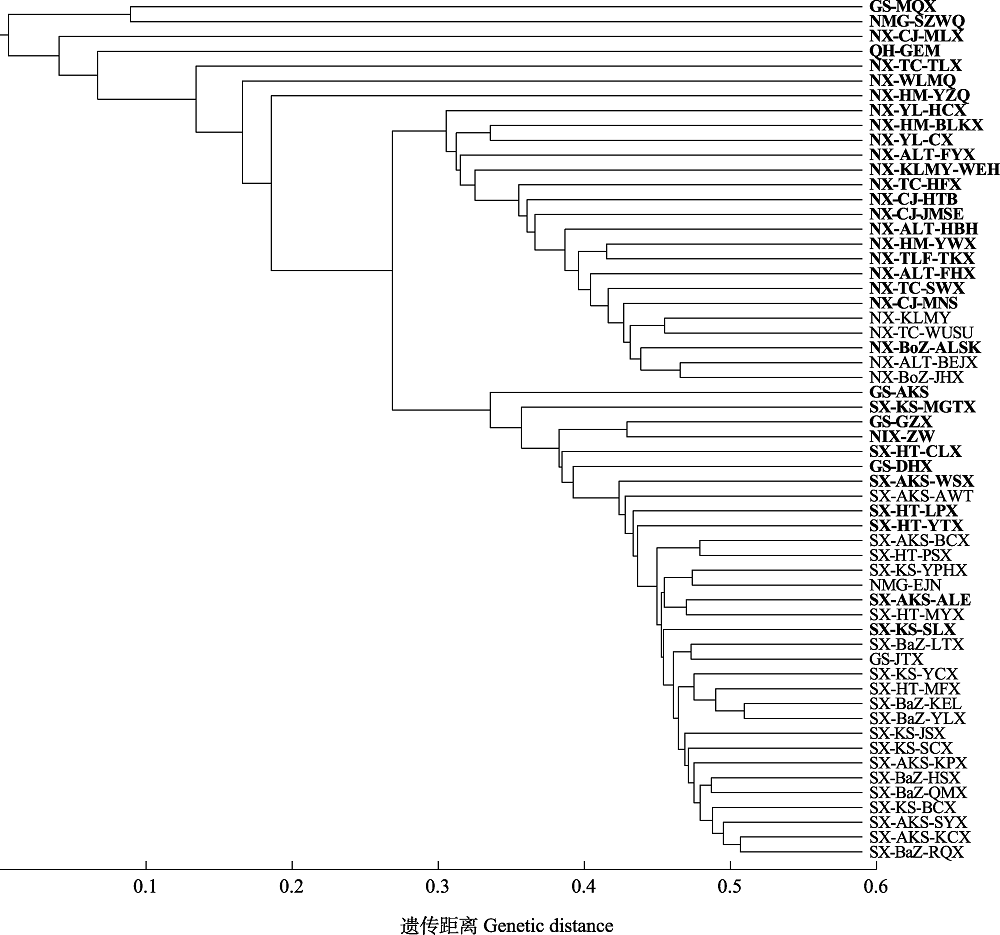
Fig. 3 The cluster analysis based on genetic distance of 58 Populus euphratica populations. See Table 1 for sampling information of the populations, and bold font indicate the 33 conservation units (CU33).
| 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC | 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC |
|---|---|---|---|---|---|---|---|---|---|
| KS-MGTX | 0.274 | 0.268 | 0.439 | 0.236 | AKS-WSX | 0.221 | 0.256 | 0.331 | 0.176 |
| KS-BCX | 0.257 | 0.265 | 0.390 | 0.207 | BaZ-YLX | 0.218 | 0.247 | 0.326 | 0.174 |
| KS-YPHX | 0.252 | 0.277 | 0.377 | 0.201 | BaZ-QMX | 0.217 | 0.254 | 0.325 | 0.173 |
| AKS-SYX | 0.246 | 0.249 | 0.376 | 0.199 | BoZ-JHX | 0.212 | 0.231 | 0.308 | 0.164 |
| AKS-BCX | 0.244 | 0.250 | 0.373 | 0.197 | HT-MFX | 0.200 | 0.266 | 0.303 | 0.161 |
| HM-YWX | 0.236 | 0.258 | 0.354 | 0.189 | TC-WUSU | 0.198 | 0.240 | 0.286 | 0.152 |
| BaZ-RQX | 0.233 | 0.248 | 0.354 | 0.187 | BaZ-HSX | 0.181 | 0.259 | 0.261 | 0.140 |
| HT-PSX | 0.231 | 0.263 | 0.351 | 0.186 | TC-HFX | 0.175 | 0.229 | 0.262 | 0.139 |
| BaZ-KEL | 0.230 | 0.255 | 0.344 | 0.183 | KLMY-WEH | 0.167 | 0.258 | 0.253 | 0.134 |
| AKS-KCX | 0.225 | 0.243 | 0.339 | 0.180 | BoZ-ALSK | 0.164 | 0.221 | 0.251 | 0.133 |
| AKS-KPX | 0.224 | 0.267 | 0.329 | 0.176 | CJ-MLX | 0.144 | 0.260 | 0.211 | 0.113 |
| BaZ-LTX | 0.223 | 0.243 | 0.330 | 0.176 |
Table 4 Genetic diversity analysis of 23 ancient Populus euphratica populations. He, Expected heterozygosity; Ho, Observed heterozygosity; I, Shannon’s information index; PIC, Polymorphism information content. Population codes see Table 1.
| 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC | 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC |
|---|---|---|---|---|---|---|---|---|---|
| KS-MGTX | 0.274 | 0.268 | 0.439 | 0.236 | AKS-WSX | 0.221 | 0.256 | 0.331 | 0.176 |
| KS-BCX | 0.257 | 0.265 | 0.390 | 0.207 | BaZ-YLX | 0.218 | 0.247 | 0.326 | 0.174 |
| KS-YPHX | 0.252 | 0.277 | 0.377 | 0.201 | BaZ-QMX | 0.217 | 0.254 | 0.325 | 0.173 |
| AKS-SYX | 0.246 | 0.249 | 0.376 | 0.199 | BoZ-JHX | 0.212 | 0.231 | 0.308 | 0.164 |
| AKS-BCX | 0.244 | 0.250 | 0.373 | 0.197 | HT-MFX | 0.200 | 0.266 | 0.303 | 0.161 |
| HM-YWX | 0.236 | 0.258 | 0.354 | 0.189 | TC-WUSU | 0.198 | 0.240 | 0.286 | 0.152 |
| BaZ-RQX | 0.233 | 0.248 | 0.354 | 0.187 | BaZ-HSX | 0.181 | 0.259 | 0.261 | 0.140 |
| HT-PSX | 0.231 | 0.263 | 0.351 | 0.186 | TC-HFX | 0.175 | 0.229 | 0.262 | 0.139 |
| BaZ-KEL | 0.230 | 0.255 | 0.344 | 0.183 | KLMY-WEH | 0.167 | 0.258 | 0.253 | 0.134 |
| AKS-KCX | 0.225 | 0.243 | 0.339 | 0.180 | BoZ-ALSK | 0.164 | 0.221 | 0.251 | 0.133 |
| AKS-KPX | 0.224 | 0.267 | 0.329 | 0.176 | CJ-MLX | 0.144 | 0.260 | 0.211 | 0.113 |
| BaZ-LTX | 0.223 | 0.243 | 0.330 | 0.176 |
| [1] |
Agiannitopoulos K, Samara P, Papadopoulou E, Tsamis K, Mertzanos G, Babalis D, Lamnissou K (2020) Study on the admission levels of circulating cell-free DNA in patients with acute myocardial infarction using different quantification methods. Scandinavian Journal of Clinical and Laboratory Investigation, 80, 1-3.
DOI URL |
| [2] |
Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature Protocols, 1, 2320-2325.
PMID |
| [3] |
Anoumaa M, Yao NK, Kouam EB, Kanmegne G, Machuka E, Osama SK, Nzuki I, Kamga YB, Fonkou T, Omokolo DN (2017) Genetic diversity and core collection for potato (Solanum tuberosum L.) cultivars from Cameroon as revealed by SSR markers. American Journal of Potato Research, 94, 449-463.
DOI URL |
| [4] |
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32, 314-331.
PMID |
| [5] |
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJA (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics, 43, 491-498.
DOI PMID |
| [6] |
Dzialuk A, Chybicki I, Gout R, Mączka T, Fleischer P, Konrad H, Curtu AL, Sofletea N, Valadon A (2014) No reduction in genetic diversity of Swiss stone pine (Pinus cembra L.) in Tatra Mountains despite high fragmentation and small population size. Conservation Genetics, 15, 1433-1445.
DOI URL |
| [7] |
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing structure output and implementing the Evanno method. Conservation Genetics Resources, 4, 359-361.
DOI URL |
| [8] |
Eusemann P, Petzold A, Thevs N, Schnittler M (2013) Growth patterns and genetic structure of Populus euphratica Oliv. (Salicaceae) forests in NW China-Implications for conservation and management. Forest Ecology and Management, 297, 27-36.
DOI URL |
| [9] |
Gai ZS, Zhai JT, Chen XX, Jiao PP, Zhang SH, Sun JH, Qin R, Liu H, Wu ZH, Li ZJ (2021) Phylogeography reveals geographic and environmental factors driving genetic differentiation of Populus sect. Turanga in northwest China. Frontiers in Plant Science, 12, 705083.
DOI URL |
| [10] | Gu YY, Zhang SQ, Li XY, Li ZJ (2013) Relationship between diameter at breast height and age of endangered species Populus euphratica Oliv. Journal of Tarim University, 25(2), 66-69. (in Chinese with English abstract) |
| [ 顾亚亚, 张世卿, 李先勇, 李志军 (2013) 濒危物种胡杨胸径与树龄关系研究. 塔里木大学学报, 25(2), 66-69.] | |
| [11] |
Hamrick JL, Linhart YB, Mitton JB (1979) Relationships between life history characteristics and electrophoretically detectable genetic variation in plants. Annual Review of Ecology and Systematics, 10, 173-200.
DOI URL |
| [12] |
Jia HX, Liu GJ, Li JB, Zhang J, Sun P, Zhao ST, Zhou X, Lu MZ, Hu JJ (2020) Genome resequencing reveals demographic history and genetic architecture of seed salinity tolerance in Populus euphratica. Journal of Experimental Botany, 71, 4308-4320.
DOI URL |
| [13] |
Kaiser SA, Taylor SA, Chen N, Sillett TS, Bondra ER, Webster MS (2017) A comparative assessment of SNP and microsatellite markers for assigning parentage in a socially monogamous bird. Molecular Ecology Resources, 17, 183-193.
DOI PMID |
| [14] |
Kansu C, Kaya Z (2020) Genetic diversity of marginal populations of Populus euphratica Oliv. from highly fragmented river ecosystems. Silvae Genetica, 69, 139-151.
DOI URL |
| [15] | Li B, Gu WC (2005) Conservation genetics of Pinus bungeana-Evaluation and conservation of natural populations’ genetic diversity. Scientia Silvae Sinicae, 41(1), 57-64. (in Chinese with English abstract) |
| [ 李斌, 顾万春 (2005) 白皮松保育遗传学--天然群体遗传多样性评价与保护策略. 林业科学, 41(1), 57-64.] | |
| [16] | Li HF, Chen TY, Huang YM, Wu CR, Li YQ, Lu SQ, Chen XT (2013) Sampling strategies of sweet potato core collection based on morphological traits. Journal of Plant Genetic Resources, 14, 91-96. (in Chinese with English abstract) |
| [ 李慧峰, 陈天渊, 黄咏梅, 吴翠荣, 李彦青, 卢森权, 陈雄庭 (2013) 基于形态性状的甘薯核心种质取样策略研究. 植物遗传资源学报, 14, 91-96.] | |
| [17] | Li ZC, Zhang HL, Zeng YW, Yang ZY, Shen SQ, Sun CQ, Wang XK (2000) Study on sampling schemes of core collection of local varieties of rice in Yunnan, China. Scientia Agricultura Sinica, 33(5), 1-7. (in Chinese with English abstract) |
| [ 李自超, 张洪亮, 曾亚文, 杨忠义, 申时全, 孙传清, 王象坤 (2000) 云南地方稻种资源核心种质取样方案研究. 中国农业科学, 33(5), 1-7.] | |
| [18] |
Liu M, Hu X, Wang X, Zhang JJ, Peng XB, Hu ZG, Liu YF (2020) Constructing a core collection of the medicinal plant Angelica biserrata using genetic and metabolic data. Frontiers in Plant Science, 11, 600249.
DOI URL |
| [19] | Ma T, Wang K, Hu QJ, Xi ZX, Wan DS, Wang Q, Feng JJ, Jiang DC, Ahani H, Abbott RJ, Lascoux M, Nevo E, Liu JQ (2018) Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proceedings of the National Academy of Sciences, USA, 115, 236-243. |
| [20] | Nagamitsu T, Shimada KI, Kanazashi A (2014) A reciprocal transplant trial suggests a disadvantage of northward seed transfer in survival and growth of Japanese red pine (Pinus densiflora) trees. Tree Genetics & Genomes, 11, 1-10. |
| [21] |
Nei M (1972) Genetic distance between populations. The American Naturalist, 106, 283-292.
DOI URL |
| [22] | Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, USA, 70, 3321-3323. |
| [23] |
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-An update. Bioinformatics, 28, 2537-2539.
PMID |
| [24] | Rohlf FJ (1998) NTSYSpc Version 2.0:User Guide. Applied Biostatistics Inc. Setauket, New York. |
| [25] | Sáenz-romero C, Snively AE, Lindig-Cisneros R (2003) Conservation and restoration of pine forest genetic resources in Mexico. Silvae Genetica, 52, 233-237. |
| [26] |
Saito Y, Shiraishi S, Tanimoto T, Yin L, Watanabe S, Ide Y (2002) Genetic diversity of Populus euphratica populations in northwestern China determined by RAPD DNA analysis. New Forests, 23, 97-103.
DOI URL |
| [27] | Shannon CE, Weaver W (1959) The Mathematical Theory of Communication. University of Illinois Press, Urbana. |
| [28] |
Tijerino A, Korpelainen H (2014) Molecular characterization of Nicaraguan Pinus tecunumanii Schw. ex Eguiluz et Perry populations for in situ conservation. Trees, 28, 1249-1253.
DOI URL |
| [29] |
Wang J, Källman T, Liu J, Guo Q, Wu Y, Lin K, Lascoux M (2014) Speciation of two desert poplar species triggered by Pleistocene climatic oscillations. Heredity, 112, 156-164.
DOI PMID |
| [30] |
Wang J, Wu YX, Ren GP, Guo QH, Liu JQ, Lascoux M (2011) Genetic differentiation and delimitation between ecologically diverged Populus euphratica and P. pruinosa. PLoS ONE, 6, e26530.
DOI URL |
| [31] | Wang SJ, Chen BH, Li HQ (1995) Populus euphratica Forest. China Environmental Science Press, Beijing. (in Chinese) |
| [ 王世绩, 陈炳浩, 李护群 (1995) 胡杨林. 中国环境科学出版社, 北京.] | |
| [32] |
Wang XL, Cao ZL, Gao CJ, Li K (2021) Strategy for the construction of a core collection for Pinus yunnanensis Franch. to optimize timber based on combined phenotype and molecular marker data. Genetic Resources and Crop Evolution, 68, 3219-3240.
DOI URL |
| [33] |
Wu YX, Wang J, Liu JQ (2008) Development and characterization of microsatellite markers in Populus euphratica (Populaceae). Molecular Ecology Resources, 8, 1142-1144.
DOI URL |
| [34] |
Xu F, Feng SS, Wu RL, Du FK (2013) Two highly validated SSR multiplexes (8-plex) for Euphrates’ poplar, Populus euphratica (Salicaceae). Molecular Ecology Resources, 13, 144-153.
DOI URL |
| [35] |
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics, 88, 76-82.
DOI URL |
| [36] | Zeng XJ, Li D, Hu YP, Huang QJ, Su XH (2014) A preliminary study on construction of high-quality core collection of Populus nigra. Scientia Silvae Sinicae, 50(9), 51-58. (in Chinese with English abstract) |
| [ 曾宪君, 李丹, 胡彦鹏, 黄秦军, 苏晓华 (2014) 欧洲黑杨优质核心种质库的初步构建. 林业科学, 50(9), 51-58.] | |
| [37] | Zeng YF, Zhang JG, Abuduhamiti B, Wang WT, Jia ZQ (2018) Phylogeographic patterns of the desert poplar in Northwest China shaped by both geology and climatic oscillations. BMC Ecology and Evolution, 18, 75. |
| [38] | Zhang L, Jiao PP, Li ZJ (2012) Genetic diversity of Populus pruinosa populations in Xinjiang of China based on SSR analysis. Chinese Journal of Ecology, 31, 2755-2761. (in Chinese with English abstract) |
| [ 张玲, 焦培培, 李志军 (2012) 中国新疆灰叶胡杨群体遗传多样性的SSR分析. 生态学杂志, 31, 2755-2761.] | |
| [39] | Zhang XC, Xu CC, Song J, Li XF (2020) Analysis of drought characteristics in Xinjiang area based on remote sensing DSI index. Jiangsu Agricultural Sciences, 48, 239-246. (in Chinese with English abstract) |
| [ 张喜成, 徐长春, 宋佳, 李晓菲 (2020) 基于遥感DSI的新疆干旱特征分析. 江苏农业科学, 48, 239-246.] | |
| [40] | Zhao B, Zhang QX (2007) Preliminary construction of the core germplasm of Chimonanthus praecox in China. Journal of Beijing Forestry University, 29(S1), 16-21. (in Chinese with English abstract) |
| [ 赵冰, 张启翔 (2007) 中国蜡梅种质资源核心种质的初步构建. 北京林业大学学报, 29(S1), 16-21.] |
| [1] | Yuanyuan Li, Chaonan Liu, Rong Wang, Shuixing Luo, Shouqian Nong, Jingwen Wang, Xiaoyong Chen. Applications of molecular markers in conserving endangered species [J]. Biodiv Sci, 2020, 28(3): 367-375. |
| [2] | Manling Wu, Lan Yao, Xunru Ai, Jiang Zhu, Qiang Zhu, Jin Wang, Xiao Huang, Jianfeng Hong. The reproductive characteristics of core germplasm in a native Metasequoia glyptostroboides population [J]. Biodiv Sci, 2020, 28(3): 303-313. |
| [3] | Yahong Zhang, Huixia Jia, Zhibin Wang, Pei Sun, Demei Cao, Jianjun Hu. Genetic diversity and population structure of Populus yunnanensis [J]. Biodiv Sci, 2019, 27(4): 355-365. |
| [4] | Jian Zhang, Lu Dong, Yanyun Zhang. Population genetic structure of Adélie penguins (Pygoscelis adeliae) from Inexpressible Island, Antarctica, using SNP markers [J]. Biodiv Sci, 2019, 27(12): 1291-1297. |
| [5] | Jie Yang, Jia He, Danbi Wang, En Shi, Wenyu Yang, Qifang Geng, Zhongsheng Wang*. Progress in research and application of InDel markers [J]. Biodiv Sci, 2016, 24(2): 237-243. |
| [6] | Dangni Zhang, Lianming Zheng, Jinru He, Wenjing Zhang, Yuanshao Lin, Yang Li. DNA barcoding of hydromedusae in northern Beibu Gulf for species identification [J]. Biodiv Sci, 2015, 23(1): 50-60. |
| [7] | Wei Zhou,Hong Wang. Pollen dispersal analysis using DNA markers [J]. Biodiv Sci, 2014, 22(1): 97-108. |
| [8] | Xueqin Wu,Gangbiao Xu,Yan Liang,Xiangbao Shen. Genetic diversity of natural and planted populations of Tsoongiodendron odorum from the Nanling Mountains [J]. Biodiv Sci, 2013, 21(1): 71-79. |
| [9] | Wen Liu, Hong Liang, Biren Yang, Jiewen Lin, Yanji Hu. Short RNAs, potential novel molecular markers for higher plants [J]. Biodiv Sci, 2012, 20(1): 86-93. |
| [10] | Zhaoxia Cui, Huan Zhang, Linsheng Song, Feng You. Genetic diversity of marine animals in China: a summary and prospec- tiveness [J]. Biodiv Sci, 2011, 19(6): 815-833. |
| [11] | Lei Li, Tong Liu, Bin Liu, Zhongquan Liu, Langming Si, Rong Zhang. Phenotypic variation and covariation among natural populations of Arabidopsis thaliana in North Xinjiang [J]. Biodiv Sci, 2010, 18(5): 497-508. |
| [12] | Ruili Zhang, Yin Jia, Qixiang Zhang. Phenotypic variation of natural populations of Primula denticulatassp. sinodenticulata [J]. Biodiv Sci, 2008, 16(4): 362-368. |
| [13] | Huifang Wu, Zuozhou Li, Hongwen Huang. Genetic differentiation among natural populations of Gastrodia elata (Orchidaceae) in Hubei and germplasm assessment of the cultivated populations [J]. Biodiv Sci, 2006, 14(4): 315-326. |
| [14] | LI Bin, GU Wan-Chun. A study on phenotypic diversity of seeds and cones characteristics in Pinus bungeana [J]. Biodiv Sci, 2002, 10(2): 181-188. |
| [15] | QIU Fang, FU Jian-Min, JIN De-Min, WANG Bin. The molecular detection of genetic diversity [J]. Biodiv Sci, 1998, 06(2): 143-150. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()