



生物多样性 ›› 2016, Vol. 24 ›› Issue (12): 1325-1334. DOI: 10.17520/biods.2016231 cstr: 32101.14.biods.2016231
所属专题: 数据论文
刘莉1,3, 舒江平1,3, 韦宏金1, 张锐1, 沈慧1,*( ), 严岳鸿1,2
), 严岳鸿1,2
收稿日期:2016-08-23
接受日期:2016-11-09
出版日期:2016-12-20
发布日期:2017-01-10
通讯作者:
沈慧
作者简介:* 通讯作者 E-mail: shenhui@sibs.ac.cn
基金资助:
Li Liu1,3, Jiangping Shu1,3, Hongjin Wei1, Rui Zhang1, Hui Shen1,*( ), Yuehong Yan1,2
), Yuehong Yan1,2
Received:2016-08-23
Accepted:2016-11-09
Online:2016-12-20
Published:2017-01-10
Contact:
Hui Shen
摘要:
岩穴蕨(Monachosorum maximowiczii)隶属于碗蕨科稀子蕨属, 是东亚中高海拔地区所特有的珍稀濒危植物。为了在分子水平对该物种有进一步的认识, 本文首次利用二代高通量测序技术(RNA-seq)对岩穴蕨进行了转录组测序分析。通过Illumina Hiseq 2500测序平台, 共计获得4.95 Gb原始数据(raw data), 经过滤后得到4.83 Gb有效数据(clean reads), 并进行从头组装得到了101,448条unigene。其中, 54,106条unigene预测到完整的开放阅读框。我们利用目前已知的51个植物基因组数据, 对岩穴蕨的unigene进行了详尽的功能注释, 并通过GO、COG、KEGG注释进一步了解了这些编码基因的作用方式、特征以及所参与的代谢通路。同时, 转录因子分析结果也为岩穴蕨的环境适应机制研究提供了初步线索。
刘莉, 舒江平, 韦宏金, 张锐, 沈慧, 严岳鸿 (2016) 东亚特有珍稀蕨类植物岩穴蕨(碗蕨科)高通量转录组测序及分析. 生物多样性, 24, 1325-1334. DOI: 10.17520/biods.2016231.
Li Liu, Jiangping Shu, Hongjin Wei, Rui Zhang, Hui Shen, Yuehong Yan (2016) De novo transcriptome analysis of the rare fern Monachosorum maximowiczii (Dennstaedtiaceae) endemic to East Asia. Biodiversity Science, 24, 1325-1334. DOI: 10.17520/biods.2016231.
| 物种名 Species | 测序部位 Description | 测序平台 Platform | 数据总量 Total size (Mb) | 单基因簇或转录本数目 No. of unigene or transcript | 文献 References |
|---|---|---|---|---|---|
| 欧洲蕨 Pteridium aquilinum | 配子体 Gametophyte | Roche 454 | 254 | 56,256 | |
| 水蕨 Ceratopteris richardii | 孢子 Spore | Roche 454 | 266 | 15,730 | |
| 鸟巢蕨 Asplenium nidus | 叶片 Leaf | Illumina HiSeq 2000 | 5,910 | 42,907 | |
| 海金沙 Lygodium japonicum | 原叶体、营养叶、孢子体、根状茎 Pro- thalli, trophophylls, sporophylls, rhizomes | Roche 454 Illumina HiSeq 2000 | 28 268 | 18,999 381,814 | |
| 巨木贼 Equisetum giganteum | 茎、叶、孢子囊 Stem, leaf, strobili | Illumina RNA-Seq | 8,800 | 34,282 |
表1 已发表的5个蕨类植物转录组测序及组装信息
Table 1 Sequencing and assembly information of five published ferns transcriptomes
| 物种名 Species | 测序部位 Description | 测序平台 Platform | 数据总量 Total size (Mb) | 单基因簇或转录本数目 No. of unigene or transcript | 文献 References |
|---|---|---|---|---|---|
| 欧洲蕨 Pteridium aquilinum | 配子体 Gametophyte | Roche 454 | 254 | 56,256 | |
| 水蕨 Ceratopteris richardii | 孢子 Spore | Roche 454 | 266 | 15,730 | |
| 鸟巢蕨 Asplenium nidus | 叶片 Leaf | Illumina HiSeq 2000 | 5,910 | 42,907 | |
| 海金沙 Lygodium japonicum | 原叶体、营养叶、孢子体、根状茎 Pro- thalli, trophophylls, sporophylls, rhizomes | Roche 454 Illumina HiSeq 2000 | 28 268 | 18,999 381,814 | |
| 巨木贼 Equisetum giganteum | 茎、叶、孢子囊 Stem, leaf, strobili | Illumina RNA-Seq | 8,800 | 34,282 |
| 原始数据 Raw data | 有效数据 Clean data | |
|---|---|---|
| 序列数目 Number of sequences | 49,027,722 | 48,497,004 |
| 全长 Total length (Gb) | 4.95 | 4.83 |
| Q20值 Q20% | 96.54% | 98.61% |
| Q30值 Q30% | 91.54% | 93.58% |
| GC值 GC% | 47.82% | 47.71% |
表2 岩穴蕨转录组测序及质控结果统计
Table 2 The results and qualities of sequencing for Monachosorum maximowiczii transcriptome
| 原始数据 Raw data | 有效数据 Clean data | |
|---|---|---|
| 序列数目 Number of sequences | 49,027,722 | 48,497,004 |
| 全长 Total length (Gb) | 4.95 | 4.83 |
| Q20值 Q20% | 96.54% | 98.61% |
| Q30值 Q30% | 91.54% | 93.58% |
| GC值 GC% | 47.82% | 47.71% |
| 转录本 Transcript | 单基因簇 Unigene | |
|---|---|---|
| 序列数目 Sequence number | 107,197 | 101,448 |
| 平均长度 Mean length (bp) | 955 | 900 |
| 最大长度 Max. length (bp) | 14,804 | 14,804 |
| 最小长度 Min. length (bp) | 201 | 201 |
| N50值 N50 | 1,907 | 1,817 |
| GC含量 GC content | 0.44 | 0.44 |
表3 岩穴蕨转录组de novo组装结果统计
Table 3 Summary of de novo assembly analysis for Monachosorum maximowiczii transcriptome
| 转录本 Transcript | 单基因簇 Unigene | |
|---|---|---|
| 序列数目 Sequence number | 107,197 | 101,448 |
| 平均长度 Mean length (bp) | 955 | 900 |
| 最大长度 Max. length (bp) | 14,804 | 14,804 |
| 最小长度 Min. length (bp) | 201 | 201 |
| N50值 N50 | 1,907 | 1,817 |
| GC含量 GC content | 0.44 | 0.44 |
| 三大类 Three categories | GO条目 GO term | Unigene 数目 No. of unigene |
|---|---|---|
| 生物学过程 Biological process | 生物过程调控 Regulation of biological process | 1,368 |
| 细胞组织部分或生物合成 Cellular component organization or biogenesis | 561 | |
| 定位活性 Establishment of localization | 1,559 | |
| 免疫系统进程 Immune system process | 9 | |
| 单一的生物过程 Single-organism process | 5,268 | |
| 定位 Localization | 1,576 | |
| 细胞活动 Locomotion | 8 | |
| 生物粘附 Biological adhesion | 2 | |
| 刺激反应 Response to stimulus | 994 | |
| 细胞杀伤 Cell killing | 1 | |
| 繁殖进程 Reproductive process | 24 | |
| 生物调节 Biological regulation | 1,448 | |
| 代谢进程 Metabolic process | 9,458 | |
| 信号传导 Signaling | 393 | |
| 生长 Growth | 16 | |
| 细胞进程 Cellular process | 8,525 | |
| 发育进程 Developmental process | 37 | |
| 有机体进程 Multi-organism process | 32 | |
| 繁殖 Reproduction | 12 | |
| 生物过程的负调控 Negative regulation of biological process | 48 | |
| 多细胞进程 Multicellular organismal process | 54 | |
| 生物过程的正调控 Positive regulation of biological process | 35 | |
| 细胞组分 Cellular component | 细胞器部分 Organelle part | 864 |
| 细胞 Cell | 2,598 | |
| 高分子复合物 Macromolecular complex | 1,399 | |
| 膜结构 Membrane | 2,329 | |
| 胞外区域 Extracellular region | 21 | |
| 膜部分 Membrane part | 1,361 | |
| 病毒体 Virion | 1 | |
| 膜关闭内腔 Membrane-enclosed lumen | 150 | |
| 细胞部分 Cell part | 2,598 | |
| 细胞器 Organelle | 1,859 | |
| 胞外基质部分 Extracellular region part | 1 | |
| 胞外基质 Extracellular matrix | 1 | |
| 病毒体部分 Virion part | 1 | |
| 分子功能 Molecular function | 结合蛋白转录活性 Protein binding transcription factor activity | 60 |
| 鸟嘌呤核苷酸交换因子活性 Guanyl-nucleotide exchange factor activity | 33 | |
| 抗氧化活性 Antioxidant activity | 114 | |
| 受体活性 Receptor activity | 105 | |
| 结构分子活性 Structural molecule activity | 294 | |
| 分子转导活性 Molecular transducer activity | 116 | |
| 酶调节活性 Enzyme regulator activity | 118 | |
| 营养库活性 Nutrient reservoir activity | 7 | |
| 金属伴侣蛋白活性 Metallochaperone activity | 4 | |
| 催化活性 Catalytic activity | 8,722 | |
| 转运活性 Transporter activity | 936 | |
| 结合活性 Binding | 9,666 | |
| 电子载体活性 Electron carrier activity | 119 | |
| 核苷酸结合转录因子活性 Nucleic acid binding transcription factor activity | 350 |
表4 岩穴蕨unigene的GO功能类别
Table 4 GO-slim functional categories of the assembled unigene for Monachosorum maximowiczii
| 三大类 Three categories | GO条目 GO term | Unigene 数目 No. of unigene |
|---|---|---|
| 生物学过程 Biological process | 生物过程调控 Regulation of biological process | 1,368 |
| 细胞组织部分或生物合成 Cellular component organization or biogenesis | 561 | |
| 定位活性 Establishment of localization | 1,559 | |
| 免疫系统进程 Immune system process | 9 | |
| 单一的生物过程 Single-organism process | 5,268 | |
| 定位 Localization | 1,576 | |
| 细胞活动 Locomotion | 8 | |
| 生物粘附 Biological adhesion | 2 | |
| 刺激反应 Response to stimulus | 994 | |
| 细胞杀伤 Cell killing | 1 | |
| 繁殖进程 Reproductive process | 24 | |
| 生物调节 Biological regulation | 1,448 | |
| 代谢进程 Metabolic process | 9,458 | |
| 信号传导 Signaling | 393 | |
| 生长 Growth | 16 | |
| 细胞进程 Cellular process | 8,525 | |
| 发育进程 Developmental process | 37 | |
| 有机体进程 Multi-organism process | 32 | |
| 繁殖 Reproduction | 12 | |
| 生物过程的负调控 Negative regulation of biological process | 48 | |
| 多细胞进程 Multicellular organismal process | 54 | |
| 生物过程的正调控 Positive regulation of biological process | 35 | |
| 细胞组分 Cellular component | 细胞器部分 Organelle part | 864 |
| 细胞 Cell | 2,598 | |
| 高分子复合物 Macromolecular complex | 1,399 | |
| 膜结构 Membrane | 2,329 | |
| 胞外区域 Extracellular region | 21 | |
| 膜部分 Membrane part | 1,361 | |
| 病毒体 Virion | 1 | |
| 膜关闭内腔 Membrane-enclosed lumen | 150 | |
| 细胞部分 Cell part | 2,598 | |
| 细胞器 Organelle | 1,859 | |
| 胞外基质部分 Extracellular region part | 1 | |
| 胞外基质 Extracellular matrix | 1 | |
| 病毒体部分 Virion part | 1 | |
| 分子功能 Molecular function | 结合蛋白转录活性 Protein binding transcription factor activity | 60 |
| 鸟嘌呤核苷酸交换因子活性 Guanyl-nucleotide exchange factor activity | 33 | |
| 抗氧化活性 Antioxidant activity | 114 | |
| 受体活性 Receptor activity | 105 | |
| 结构分子活性 Structural molecule activity | 294 | |
| 分子转导活性 Molecular transducer activity | 116 | |
| 酶调节活性 Enzyme regulator activity | 118 | |
| 营养库活性 Nutrient reservoir activity | 7 | |
| 金属伴侣蛋白活性 Metallochaperone activity | 4 | |
| 催化活性 Catalytic activity | 8,722 | |
| 转运活性 Transporter activity | 936 | |
| 结合活性 Binding | 9,666 | |
| 电子载体活性 Electron carrier activity | 119 | |
| 核苷酸结合转录因子活性 Nucleic acid binding transcription factor activity | 350 |
| COG功能分类 COG function classifications | Unigene数目 No. of unigene | 百分比 Percentage (%) |
|---|---|---|
| RNA加工与修饰 RNA processing and modification | 576 | 4.76 |
| 染色质结构和活力 Chromatin structure and dynamics | 168 | 1.39 |
| 能量生成与转换 Energy production and conversion | 475 | 3.92 |
| 细胞周期控制、细胞分裂、染色体分区 Cell cycle control, cell division, chromosome partitioning | 288 | 2.38 |
| 氨基酸转运与代谢 Amino acid transport and metabolism | 318 | 2.63 |
| 核苷酸转运与代谢 Nucleotide transport and metabolism | 181 | 1.49 |
| 碳水化合物转运与代谢 Carbohydrate transport and metabolism | 546 | 4.51 |
| 辅酶转运与代谢 Coenzyme transport and metabolism | 154 | 1.27 |
| 脂类转运与代谢 Lipid transport and metabolism | 424 | 3.50 |
| 翻译、核糖体结构和生物发生 Translation, ribosomal structure and biogenesis | 562 | 4.64 |
| 转录 Transcription | 669 | 5.52 |
| 复制、重组和修复 Replication, recombination and repair | 403 | 3.33 |
| 细胞壁膜生物合成 Cell wall membrane biogenesis | 66 | 0.54 |
| 细胞运动 Cell motility | 2 | 0.02 |
| 翻译后修饰、蛋白质折叠、分子伴侣 Posttranslational modification, protein turnover, chaperones | 1,209 | 9.98 |
| 无机离子转运与代谢 Inorganic ion transport and metabolism | 315 | 2.60 |
| 次生代谢物合成、转运与代谢 Secondary metabolites biosynthesis, transport and catabolism | 441 | 3.64 |
| 一般功能预测 General function prediction only | 1,380 | 11.39 |
| 未知功能 Function unknown | 932 | 7.69 |
| 信号转导机制 Signal transduction mechanisms | 1,933 | 15.96 |
| 细胞内转运、分泌和小泡运输 Intracellular trafficking, secretion, and vesicular transport | 610 | 5.04 |
| 防卫机制 Defense mechanisms | 114 | 0.94 |
| 胞外结构 Extracellular structures | 56 | 0.46 |
| 核结构 Nuclear structure | 32 | 0.26 |
| 细胞骨架 Cytoskeleton | 259 | 2.14 |
表5 岩穴蕨unigene的COG分类
Table 5 Classification of the clusters of orthologous groups (COG) for Monachosorum maximowiczii
| COG功能分类 COG function classifications | Unigene数目 No. of unigene | 百分比 Percentage (%) |
|---|---|---|
| RNA加工与修饰 RNA processing and modification | 576 | 4.76 |
| 染色质结构和活力 Chromatin structure and dynamics | 168 | 1.39 |
| 能量生成与转换 Energy production and conversion | 475 | 3.92 |
| 细胞周期控制、细胞分裂、染色体分区 Cell cycle control, cell division, chromosome partitioning | 288 | 2.38 |
| 氨基酸转运与代谢 Amino acid transport and metabolism | 318 | 2.63 |
| 核苷酸转运与代谢 Nucleotide transport and metabolism | 181 | 1.49 |
| 碳水化合物转运与代谢 Carbohydrate transport and metabolism | 546 | 4.51 |
| 辅酶转运与代谢 Coenzyme transport and metabolism | 154 | 1.27 |
| 脂类转运与代谢 Lipid transport and metabolism | 424 | 3.50 |
| 翻译、核糖体结构和生物发生 Translation, ribosomal structure and biogenesis | 562 | 4.64 |
| 转录 Transcription | 669 | 5.52 |
| 复制、重组和修复 Replication, recombination and repair | 403 | 3.33 |
| 细胞壁膜生物合成 Cell wall membrane biogenesis | 66 | 0.54 |
| 细胞运动 Cell motility | 2 | 0.02 |
| 翻译后修饰、蛋白质折叠、分子伴侣 Posttranslational modification, protein turnover, chaperones | 1,209 | 9.98 |
| 无机离子转运与代谢 Inorganic ion transport and metabolism | 315 | 2.60 |
| 次生代谢物合成、转运与代谢 Secondary metabolites biosynthesis, transport and catabolism | 441 | 3.64 |
| 一般功能预测 General function prediction only | 1,380 | 11.39 |
| 未知功能 Function unknown | 932 | 7.69 |
| 信号转导机制 Signal transduction mechanisms | 1,933 | 15.96 |
| 细胞内转运、分泌和小泡运输 Intracellular trafficking, secretion, and vesicular transport | 610 | 5.04 |
| 防卫机制 Defense mechanisms | 114 | 0.94 |
| 胞外结构 Extracellular structures | 56 | 0.46 |
| 核结构 Nuclear structure | 32 | 0.26 |
| 细胞骨架 Cytoskeleton | 259 | 2.14 |
| 类别 Categories | 通路 Pathways | Unigene数目 No. of unigene | 通路数 No. of pathways |
|---|---|---|---|
| 代谢途径 Metabolism etabolism | 碳水化合物代谢 Carbohydrate metabolism | 1,511 | 15 |
| 脂类物质代谢 Lipid metabolism | 813 | 14 | |
| 辅助因子和维生素代谢 Metabolism of cofactors and vitamins | 371 | 11 | |
| 能量代谢 Energy metabolism | 477 | 6 | |
| 核苷酸代谢 Nucleotide metabolism | 446 | 2 | |
| 氨基酸代谢 Amino acid metabolism | 831 | 14 | |
| 多酮类和萜类化合物的代谢 Metabolism of terpenoids and polyketides | 224 | 8 | |
| 其他次生代谢产物的生物合成 Biosynthesis of other secondary metabolites | 314 | 8 | |
| 其他氨基酸代谢 Metabolism of other amino acids | 270 | 5 | |
| 多糖生物合成与代谢 Glycan biosynthesis and metabolism | 166 | 7 | |
| 全球与总体图 Global and overview maps 小计 Sub-total | 4,956 10,379 (73.41%) | 6 96 (75%) | |
| 遗传信息加工 Genetic information processing | 折叠、分类和降解 Folding, sorting and degradation | 769 | 7 |
| 翻译 Translation | 876 | 5 | |
| 转录 Transcription | 413 | 3 | |
| 复制与修复 Replication and repair 小计 Sub-total | 493 2,551 (18.04%) | 6 21 (16.41%) | |
| 环境信息处理 Environmental information processing | 信号转导 Signal transduction 膜运输 Membrane transport 小计 Sub-total | 350 36 386 (2.73%) | 2 1 3 (2.34%) |
| 细胞过程 Cellular processes | 运输与代谢 Transport and catabolism 小计 Sub-total | 506 506 (3.58%) | 4 4 (3.13%) |
| 生物系统 Organismal systems | 环境适应性 Environmental adaptation 小计 Sub-total | 252 252 (1.78%) | 2 2 (1.56%) |
| 人类疾病 Human diseases | 耐药性: 抗菌 Drug resistance: Antimicrobial | 7 | 1 |
| 内分泌与代谢性疾病 Endocrine and metabolic diseases 小计 Sub-total | 57 64 (0.45%) | 1 2 (1.56%) |
表6 岩穴蕨unigene的KEGG注释结果及分类
Table 6 The KEGG classification and metabolism pathways for the annotated unigene for Monachosorum maximowiczii
| 类别 Categories | 通路 Pathways | Unigene数目 No. of unigene | 通路数 No. of pathways |
|---|---|---|---|
| 代谢途径 Metabolism etabolism | 碳水化合物代谢 Carbohydrate metabolism | 1,511 | 15 |
| 脂类物质代谢 Lipid metabolism | 813 | 14 | |
| 辅助因子和维生素代谢 Metabolism of cofactors and vitamins | 371 | 11 | |
| 能量代谢 Energy metabolism | 477 | 6 | |
| 核苷酸代谢 Nucleotide metabolism | 446 | 2 | |
| 氨基酸代谢 Amino acid metabolism | 831 | 14 | |
| 多酮类和萜类化合物的代谢 Metabolism of terpenoids and polyketides | 224 | 8 | |
| 其他次生代谢产物的生物合成 Biosynthesis of other secondary metabolites | 314 | 8 | |
| 其他氨基酸代谢 Metabolism of other amino acids | 270 | 5 | |
| 多糖生物合成与代谢 Glycan biosynthesis and metabolism | 166 | 7 | |
| 全球与总体图 Global and overview maps 小计 Sub-total | 4,956 10,379 (73.41%) | 6 96 (75%) | |
| 遗传信息加工 Genetic information processing | 折叠、分类和降解 Folding, sorting and degradation | 769 | 7 |
| 翻译 Translation | 876 | 5 | |
| 转录 Transcription | 413 | 3 | |
| 复制与修复 Replication and repair 小计 Sub-total | 493 2,551 (18.04%) | 6 21 (16.41%) | |
| 环境信息处理 Environmental information processing | 信号转导 Signal transduction 膜运输 Membrane transport 小计 Sub-total | 350 36 386 (2.73%) | 2 1 3 (2.34%) |
| 细胞过程 Cellular processes | 运输与代谢 Transport and catabolism 小计 Sub-total | 506 506 (3.58%) | 4 4 (3.13%) |
| 生物系统 Organismal systems | 环境适应性 Environmental adaptation 小计 Sub-total | 252 252 (1.78%) | 2 2 (1.56%) |
| 人类疾病 Human diseases | 耐药性: 抗菌 Drug resistance: Antimicrobial | 7 | 1 |
| 内分泌与代谢性疾病 Endocrine and metabolic diseases 小计 Sub-total | 57 64 (0.45%) | 1 2 (1.56%) |
| 编码 ID | 代谢通路名称 Pathway | unigene数目 No. of unigene | 百分比 Percentage (%) |
|---|---|---|---|
| ko01100 | 代谢通路 Metabolic pathways | 2,618 | 23.03 |
| ko01110 | 次生代谢产物的生物合成 Biosynthesis of secondary metabolites | 1,418 | 12.47 |
| ko01200 | 碳代谢 Carbon metabolism | 406 | 3.57 |
| ko01230 | 氨基酸生物合成 Biosynthesis of amino acids | 330 | 2.90 |
| ko03040 | 剪接体 Spliceosome | 297 | 2.61 |
| ko03010 | 核糖体 Ribosome | 292 | 2.57 |
| ko00500 | 淀粉与蔗糖代谢 Starch and sucrose metabolism | 263 | 2.31 |
| ko04075 | 植物激素信号转导 Plant hormone signal transduction | 255 | 2.24 |
| ko00230 | 嘌呤代谢 Purine metabolism | 243 | 2.14 |
| ko04141 | 内质网中的蛋白质加工 Protein processing in endoplasmic reticulum | 230 | 2.02 |
表7 KEGG分类中unigene数量最多的10个代谢通路
Table 7 Top ten metabolism pathways involving unigene in KEGG
| 编码 ID | 代谢通路名称 Pathway | unigene数目 No. of unigene | 百分比 Percentage (%) |
|---|---|---|---|
| ko01100 | 代谢通路 Metabolic pathways | 2,618 | 23.03 |
| ko01110 | 次生代谢产物的生物合成 Biosynthesis of secondary metabolites | 1,418 | 12.47 |
| ko01200 | 碳代谢 Carbon metabolism | 406 | 3.57 |
| ko01230 | 氨基酸生物合成 Biosynthesis of amino acids | 330 | 2.90 |
| ko03040 | 剪接体 Spliceosome | 297 | 2.61 |
| ko03010 | 核糖体 Ribosome | 292 | 2.57 |
| ko00500 | 淀粉与蔗糖代谢 Starch and sucrose metabolism | 263 | 2.31 |
| ko04075 | 植物激素信号转导 Plant hormone signal transduction | 255 | 2.24 |
| ko00230 | 嘌呤代谢 Purine metabolism | 243 | 2.14 |
| ko04141 | 内质网中的蛋白质加工 Protein processing in endoplasmic reticulum | 230 | 2.02 |
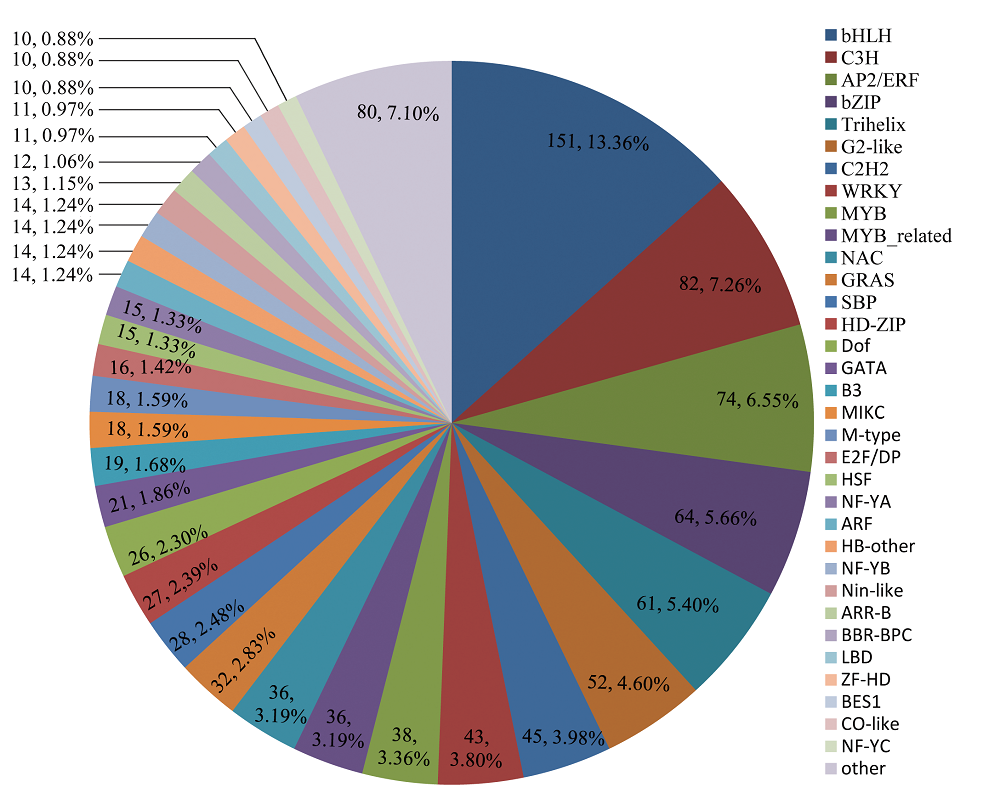
图2 岩穴蕨unigene的转录因子预测。将54,106个预测到ORF的unigene比对到拟南芥(Arabidopsis thaliana)的转录因子数据库, 预测到1,130个转录因子。
Fig. 2 Number of Monachosorum maximowiczii unigene that were predicted as transcription factors. The 54,106 unigene which had been predicted ORF were aligned to Arabidopsis thaliana in PlantTFDB v3.0, and 1,130 unigene were predicted as transcription factors.
| [1] |
Aya K, Kobayashi M, Tanaka J, Ohyanagi H, Suzuki T, Yano K, Takano T, Yano K, Matsuoka M (2015) De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database, Ljtrans DB. Plant and Cell Physiology, 56, e5.
DOI URL |
| [2] |
Bushart TJ, Cannon AE, ul Haque A, San MP, Mostajeran K, Clark GB, Porterfield DM, Roux SJ (2013) RNA-seq analysis identifies potential modulators of gravity response in spores of Ceratopteris (Parkeriaceae): evidence for modulation by calcium pumps and apyrase activity. American Journal of Botany, 100, 161-174.
DOI PMID |
| [3] |
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Development, 17, 1043-1054.
DOI URL |
| [4] | Ching RC (1978) The Chinese fern families and genera: systematic arrangement and historical origin (cont.). Acta Phytotaxonomica Sinica, 16(4), 16-37. (in Chinese) |
| [秦仁昌 (1978) 中国蕨类植物科属的系统排列和历史来源(续). 植物分类学报, 16(4), 16-37.] | |
| [5] |
Christenhusz MJM, Byng JW (2016) The number of known plants species in the world and its annual increase. Phytotaxa, 261, 201-217.
DOI URL |
| [6] | Costa V, Angelini C, de Feis I, Ciccodicola A (2010) Uncovering the complexity of transcriptomes with RNA-Seq. BioMed Research International, 2010, 853916. |
| [7] |
Der JP, Barker MS, Wickett NJ, dePamphilis CW, Wolf PG (2011) De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BioMed Central Genomics, 12, doi: 10.1186/1471-2164-12-99.
DOI |
| [8] |
Franssen SU, Shrestha RP, Brautigam A, Bornberg BE, Weber AP (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BioMed Central Genomics, 12, doi: 10.1186/1471-2164-12-227.
DOI |
| [9] | Jia CL, Zhang Y, Zhu L, Zhang R (2015) Application progress of transcriptome sequencing technology in biological sequencing. Molecular Plant Breeding, 13, 2388-2394. (in Chinese with English abstract) |
| [贾昌路, 张瑶, 朱玲, 张锐 (2015) 转录组测序技术在生物测序中的应用研究进展. 分子植物育种, 13, 2388-2394.] | |
| [10] | Jia XP, Sun XB, Deng YM, Liang LJ, Ye XQ (2014) Sequencing and analysis of the transcriptome of Asplenium nidus. Acta Horticulturae Sinica, 41, 2329-2341. (in Chinese with English abstract) |
| [贾新平, 孙晓波, 邓衍明, 梁丽建, 叶晓青 (2014) 鸟巢蕨转录组高通量测序及分析. 园艺学报, 41, 2329-2341.] | |
| [11] |
Li H, Yao WJ, Fu Y, Li S, Guo QQ (2015) De novo assembly and discovery of genes that are involved in drought tolerance in Tibetan Sophora moorcroftiana. PLoS ONE, 10, e111054.
DOI URL |
| [12] | Liu H, Li DJ, Deng Z (2014) Advances in research of transcriptional regulatory network in response to cold stress in plants. Scientia Agricultura Sinica, 47, 3523-3533. (in Chinese with English abstract) |
| [刘辉, 李德军, 邓治 (2014) 植物应答低温胁迫的转录调控网络研究进展. 中国农业科学, 47, 3523-3533.] | |
| [13] | Qi CJ, Yu XL, Cao TR, Zhou JR (1994) Flora of Hunan Badagongshan Mountains and its phytogeographical significance. Acta Botanica Yunnanica, 16, 321-332. (in Chinese with English abstract) |
| [祁承经, 喻勋林, 曹铁如, 周建仁 (1994) 湖南八大公山的植物区系及其在植物地理学上的意义. 云南植物研究, 16, 321-332.] | |
| [14] |
Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD (2011) OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. The Plant Journal, 65, 907-921.
DOI URL |
| [15] | Vannesta K, Sterck L, Mybury AA, Peer YV, Mizrachi E (2015) Horsetails are ancient polyploids: evidence from Equisetum giganteum. The Plant Cell, 27, 1567-1568. |
| [16] |
Wang Y, Jiang CJ, Li YY, Wei CL, Deng WW (2011) CsICE1 and CsCBF1: two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Reports, 31, 27-34.
DOI URL |
| [17] |
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics, 10, 57-63.
DOI URL |
| [18] | Wei LB, Miao HM, Zhang HY (2012) Transcriptomic analysis of sesame development. Scientia Agricultura Sinica, 45, 1246-1256.(in Chinese with English abstract) |
| [魏利斌, 苗红梅, 张海洋 (2012) 芝麻发育转录组分析. 中国农业科学, 45, 1246-1256.] | |
| [19] | Wu Y, Wei W, Pang XY, Wang XF, Zhang HL, Dong B, Xing YP, Li XG, Wang MY (2014) Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BioMed Central Genomics, 15, 1-16. |
| [20] | Wu ZY, Raven P, Hong DY (2013) Flora of China. Vol.2 (Dennstaedtiaceae). Science Press, Beijing; Missouri Botanical Garden Press, St. Louis. |
| [21] | Yang N, Zhao KG, Chen LQ (2012) Deep sequencing-based transcriptome profiling analysis of Chimonanthus praecox reveals insights into secondary metabolites biosynthesis. Journal of Beijing Forestry University, 34(1), 104-107. (in Chinese with English abstract) |
| [杨楠, 赵凯歌, 陈龙清 (2012) 蜡梅花转录组数据分析及次生代谢产物合成途径研究. 北京林业大学学报, 34(1), 104-107.] | |
| [22] | Yang Y, Li X, Kong X, Ma L, Hu X, Yang Y (2015) Transcriptome analysis reveals diversified adaptation of Stipa purpurea along a drought gradient on the Tibetan Plateau. Functional & Integrative Genomics, 15, 295-307. |
| [23] | Zhang LL, Li JF, Wang AX (2008) The role of the transcription factor CBF genes in cold-responsive molecular mechanism. Acta Horticulturae Sinica, 35, 765-771. (in Chinese with English abstract) |
| [张丽丽, 李景富, 王傲雪 (2008) 转录激活因子CBF基因在植物抗冷分子机制中的作用. 园艺学报, 35, 765-771.] | |
| [24] |
Zhang X, Wang JH, Yu M, Cao K, Zhuang L, Xu CX, Cao WD (2015) Transcriptome analysis of bioenergy plant Miscanthus sinensis Anderss by RNA-Seq. Chinese Journal of Biotechnology, 31, 1437-1448. (in Chinese with English abstract)
PMID |
|
[张贤, 王建红, 喻曼, 庄俐, 徐昌旭, 曹卫东 (2015) 基于RNA-Seq的能源植物芒转录组分析. 生物工程学报, 31, 1437-1448.]
PMID |
|
| [25] |
Zhou D, Gao S, Wang H, Lei T, Shen J, Gao J, Chen S (2016) De novo sequencing transcriptome of endemic Gentiana straminea (Gentianaceae) to identify genes involved in the biosynthesis of active ingredients. Gene, 575, 160-170.
DOI URL |
| [26] |
Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY (2009) Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. Journal of Plant Physiology, 166, 1296-1306.
DOI URL |
| [27] | Zhu SQ, Gong YF, Hang YQ, Liu H, Wang HY (2015) Transcriptome analysis of Dunaliella viridis. Hereditas, 37, 828-836. (in Chinese with English abstract) |
| [朱帅旗, 龚一富, 杭雨晴, 刘浩, 王何瑜 (2015) 绿色杜氏藻转录组分析. 遗传, 37, 828-836.] |
| [1] | 干靓 刘巷序 鲁雪茗 岳星. 全球生物多样性热点地区大城市的保护政策与优化方向[J]. 生物多样性, 2025, 33(5): 24529-. |
| [2] | 曾子轩 杨锐 黄越 陈路遥. 清华大学校园鸟类多样性特征与环境关联[J]. 生物多样性, 2025, 33(5): 24373-. |
| [3] | 臧明月, 刘立, 马月, 徐徐, 胡飞龙, 卢晓强, 李佳琦, 于赐刚, 刘燕. 《昆明-蒙特利尔全球生物多样性框架》下的中国城市生物多样性保护[J]. 生物多样性, 2025, 33(5): 24482-. |
| [4] | 祝晓雨, 王晨灏, 王忠君, 张玉钧. 城市绿地生物多样性研究进展与展望[J]. 生物多样性, 2025, 33(5): 25027-. |
| [5] | 袁琳, 王思琦, 侯静轩. 大都市地区的自然留野:趋势与展望[J]. 生物多样性, 2025, 33(5): 24481-. |
| [6] | 胡敏, 李彬彬, Coraline Goron. 只绿是不够的: 一个生物多样性友好的城市公园管理框架[J]. 生物多样性, 2025, 33(5): 24483-. |
| [7] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果分析[J]. 生物多样性, 2025, 33(5): 24531-. |
| [8] | 明玥, 郝培尧, 谭铃千, 郑曦. 基于城市绿色高质量发展理念的中国城市生物多样性保护与提升研究[J]. 生物多样性, 2025, 33(5): 24524-. |
| [9] | 谢淦, 宣晶, 付其迪, 魏泽, 薛凯, 雒海瑞, 高吉喜, 李敏. 草地植物多样性无人机调查的物种智能识别模型构建[J]. 生物多样性, 2025, 33(4): 24236-. |
| [10] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [11] | 宋威, 程才, 王嘉伟, 吴纪华. 土壤微生物对植物多样性–生态系统功能关系的调控作用[J]. 生物多样性, 2025, 33(4): 24579-. |
| [12] | 卢晓强, 董姗姗, 马月, 徐徐, 邱凤, 臧明月, 万雅琼, 李孪鑫, 于赐刚, 刘燕. 前沿技术在生物多样性研究中的应用现状、挑战与展望[J]. 生物多样性, 2025, 33(4): 24440-. |
| [13] | 农荞伊, 曹军, 程文达, 彭艳琼. 不同方法对蜜蜂总科昆虫资源与多样性监测效果的比较[J]. 生物多样性, 2025, 33(4): 25057-. |
| [14] | 郭雨桐, 李素萃, 王智, 解焱, 杨雪, 周广金, 尤春赫, 朱萨宁, 高吉喜. 全国自然保护地对国家重点保护野生物种的覆盖度及其分布状况[J]. 生物多样性, 2025, 33(3): 24423-. |
| [15] | 赵维洋, 王伟, 马冰然. 其他有效的区域保护措施(OECMs)研究进展与展望[J]. 生物多样性, 2025, 33(3): 24525-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn