



生物多样性 ›› 2019, Vol. 27 ›› Issue (11): 1221-1227. DOI: 10.17520/biods.2019236 cstr: 32101.14.biods.2019236
汪浩1,2#,张锐1#,张娇1,沈慧1,戴锡玲2,严岳鸿1,*( )
)
收稿日期:2019-07-25
接受日期:2019-12-17
出版日期:2019-11-20
发布日期:2020-01-17
通讯作者:
严岳鸿
基金资助:
Hao Wang1,2#,Rui Zhang1#,Jiao Zhang1,Hui Shen1,Xiling Dai2,Yuehong Yan1,*( )
)
Received:2019-07-25
Accepted:2019-12-17
Online:2019-11-20
Published:2020-01-17
Contact:
Yan Yuehong
摘要:
全基因组复制在动植物中普遍存在, 被认为是促进物种进化的重要动力之一。作为蕨类植物的单种科物种, 翼盖蕨(Didymochlaena trancatula)是真水龙骨类I的基部类群, 在蕨类中具有独特的演化地位。本研究基于高通量测序, 通过同义替换率(Ks)分析、相对定年分析揭示翼盖蕨的全基因组复制发生情况。Ks分析表明, 翼盖蕨至少经历了两次全基因组复制事件, 其中一次发生于59-62 million years ago (Mya), 另一次发生于90-94 Mya, 这两次全基因组复制事件分别和白垩纪第三纪的Cretaceous-Tertiary (C-T)大灭绝事件以及翼盖蕨的物种分化时间相吻合。进一步对两次全基因组复制保留的基因进行功能注释和富集分析, 结果显示与转录及代谢调控相关的基因优势被保留。翼盖蕨的全基因组复制事件可能促进了该物种的分化及其对极端环境的适应性。
汪浩, 张锐, 张娇, 沈慧, 戴锡玲, 严岳鸿 (2019) 转录组测序揭示翼盖蕨(Didymochlaena trancatula)的全基因组复制历史. 生物多样性, 27, 1221-1227. DOI: 10.17520/biods.2019236.
Hao Wang, Rui Zhang, Jiao Zhang, Hui Shen, Xiling Dai, Yuehong Yan (2019) De novo transcriptome assembly reveals the whole genome duplication events of Didymochlaena trancatula. Biodiversity Science, 27, 1221-1227. DOI: 10.17520/biods.2019236.
| 转录本 Transcript | 单基因簇 Unigene | |
|---|---|---|
| 序列数目 Sequence number | 77,709 | 58,871 |
| 平均长度 Mean length (bp) | 986 | 836 |
| 最大长度 Max. length (bp) | 17,374 | 17,374 |
| 最小长度 Min. length (bp) | 201 | 201 |
| N50 | 1,734 | 1,546 |
表1 翼盖蕨转录组de novo组装结果统计
Table 1 Summary of de novo assembly for Didymochlaena trancatula transcriptome
| 转录本 Transcript | 单基因簇 Unigene | |
|---|---|---|
| 序列数目 Sequence number | 77,709 | 58,871 |
| 平均长度 Mean length (bp) | 986 | 836 |
| 最大长度 Max. length (bp) | 17,374 | 17,374 |
| 最小长度 Min. length (bp) | 201 | 201 |
| N50 | 1,734 | 1,546 |
| 加倍数目 No. of duplications | 组件数目 No. of components | 贝叶斯信息标准 Bayesian information criterion | 中值 Median | 方差 Variance | 比例 Proportion |
|---|---|---|---|---|---|
| 1,951 | 7 | 5,012.931 | 0.1211 | 0.0002 | 0.0953 |
| 1,951 | 7 | 5,012.931 | 0.1796 | 0.0015 | 0.0897 |
| 1,951 | 7 | 5,012.931 | 0.3518 | 0.0134 | 0.0584 |
| 1,951 | 7 | 5,012.931 | 1.3359 | 0.1921 | 0.3296 |
| 1,951 | 7 | 5,012.931 | 2.0329 | 0.1872 | 0.2107 |
| 1,951 | 7 | 5,012.931 | 3.2182 | 0.5590 | 0.1958 |
| 1,951 | 7 | 5,012.931 | 4.7933 | 0.0105 | 0.0205 |
表2 基于混合模型拟合的翼盖蕨的Ks结果
Table 2 Ks results of Didymochlaena trancatula based on mixture modeling
| 加倍数目 No. of duplications | 组件数目 No. of components | 贝叶斯信息标准 Bayesian information criterion | 中值 Median | 方差 Variance | 比例 Proportion |
|---|---|---|---|---|---|
| 1,951 | 7 | 5,012.931 | 0.1211 | 0.0002 | 0.0953 |
| 1,951 | 7 | 5,012.931 | 0.1796 | 0.0015 | 0.0897 |
| 1,951 | 7 | 5,012.931 | 0.3518 | 0.0134 | 0.0584 |
| 1,951 | 7 | 5,012.931 | 1.3359 | 0.1921 | 0.3296 |
| 1,951 | 7 | 5,012.931 | 2.0329 | 0.1872 | 0.2107 |
| 1,951 | 7 | 5,012.931 | 3.2182 | 0.5590 | 0.1958 |
| 1,951 | 7 | 5,012.931 | 4.7933 | 0.0105 | 0.0205 |
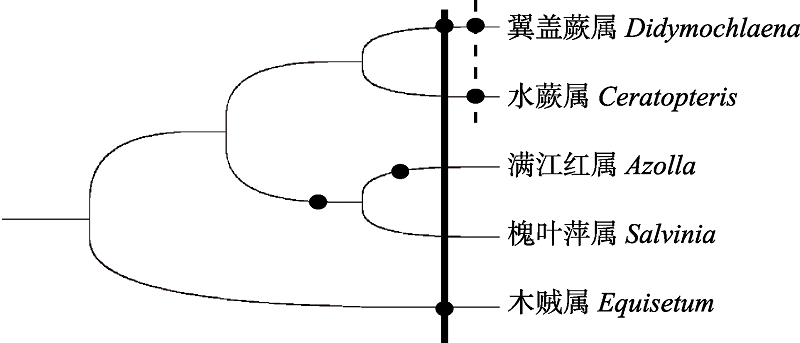
图3 蕨类中已报道的WGDs发生概况。圆圈代表WGD, 实线代表翼盖蕨和巨木贼中(约90 Mya)发生WGD的时间; 虚线代表翼盖蕨和水蕨中(约50 Mya)发生WGD的时间。
Fig. 3 WGDs occurrence in ferns from previous reports. Circle represents WGDs. The solid line denotes the age of WGD at (about 90 Mya) Didymochlaena trancatula and Equisetum giganteum, the dash line indicates the age of WGD both detected in Didymochlaena trancatula and Ceretopteris thalictroides (about 50 Mya).
| 1 | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G ( 2000) Gene ontology: Tool for the unification of biology. Nature Genetic, 25, 25-29. |
| 2 | Athanasios T, Joseph RE, Ronald WD ( 2000) Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature, 408, 816-820. |
| 3 | Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B, Legrand L, Gill N, Kane NC, Bowers JE, Hubner S, Bellec A, Bérard A, Bergès H, Blanchet N, Boniface MC, Brunel D, Catrice O, Chaidir N, Claudel C, Donnadieu C, Faraut T, Fievet G, Helmstetter N, King M, Knapp SJ, Lai Z, Le Paslier MC, Lippi Y, Lorenzon L, Mandel JR, Marage G, Marchand G, Marquand E, Bret-Mestries E, Morien E, Nambeesan S, Nguyen T, Pegot-Espagnet P, Pouilly N, Raftis F, Sallet E, Schiex T, Thomas J, Vandecasteele C, Varès D, Vear F, Vautrin S, Crespi M, Mangin B, Burke JM, Salse J, Muños S, Vincourt P, Rieseberg LH, Langlade NB ( 2017) The sunflower genome provides insights into oil metabolism, flowering and asterid evolution. Nature, 546, 148-152. |
| 4 | Barker MS ( 2009) Evolutionary genomic analyses of ferns reveal that high chromosome numbers are a product of high retention and fewer rounds of polyploidy relative to angiosperms. American Fern Journal, 99, 136-141. |
| 5 | Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA ( 2015) On the relative abundance of auto- and allopolyploids. New Phytologist, 210, 391-398. |
| 6 | Blanc G, Wolfe KH ( 2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell, 16, 1667-1678. |
| 7 | Bowers JE, Chapman BA, Rong J, Paterson AH ( 2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature, 422, 433-438. |
| 8 | Clark JW, Puttick MN, Donoghue PCJ ( 2019) Origin of horsetails and the role of whole-genome duplication in plant macroevolution. Proceedings of the Royal Society B: Biological Sciences, 286, 20191662. |
| 9 | Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, Albert VA, Ma H, dePamphilis CW ( 2006) Widespread genome duplications throughout the history of flowering plants. Genome Research, 16, 738-749. |
| 10 | De Bodt S, Maere S, Van de Peer Y ( 2005) Genome duplication and the origin of angiosperms. Trends in Ecology & Evolution, 20, 591-597. |
| 11 | Dyer RJ, Pellicer J, Savolainen V, Leitch IJ, Schneider H ( 2013) Genome size expansion and the relationship between nuclear DNA content and spore size in the Asplenium monanthes fern complex (Aspleniaceae). BMC Plant Biology, 13, 219. |
| 12 | Edgar RC ( 2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792-1797. |
| 13 | Enright AJ, Van Dongen S, Ouzounis CA ( 2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Research, 30, 1575-1584. |
| 14 | Fawcett JA, Maere S, Van de Peer Y ( 2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proceedings of the National Academy of Sciences, USA, 106, 5737-5742. |
| 15 | Fraley C, Raftery AE ( 2003) Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST. Journal of Classification, 20, 263-286. |
| 16 | Freeling M ( 2009) Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annual Review of Plant Biology, 60, 433-453. |
| 17 | Goldman N, Yang Z ( 1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution, 11, 725-736. |
| 18 | Guerra D, Crosatti C, Khoshro HH, Mastrangelo AM, Mica E, Mazzucotelli E ( 2015) Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Frontier in Plant Science, 6, 57. |
| 19 | Hegarty MJ, Hiscock SJ ( 2008) Genomic clues to the evolutionary success of polyploid plants. Current Biology, 18, 435-444. |
| 20 | Huang CH, Qi X, Chen D, Qi J, Ma H ( 2019) Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. Journal of Integrative Plant Biology, doi: 10.1111/jipb.12877. |
| 21 | Huang CH, Zhang C, Liu M, Hu Y, Gao T, Qi J, Ma H ( 2016) Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Molecular Biology and Evolution, 33, 2820-2835. |
| 22 | Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW ( 2011) Ancestral polyploidy in seed plants and angiosperms. Nature, 473, 97-100. |
| 23 | Kimura M ( 1977) Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature, 267, 275-276. |
| 24 | Li FW, Brouwer P, Carretero-Paulet L, Cheng S, de Vries J, Delaux PM, Eily A, Koppers N, Kuo LY, Li Z, Simenc M, Small I, Wafula E, Angarita S, Barker MS, Bräutigam A, dePamphilis C, Gould S, Hosmani PS, Huang YM, Huettel B, Kato Y, Liu X, Maere S, McDowell R, Mueller LA, Nierop KGJ, Rensing SA, Robison T, Rothfels CJ, Sigel EM, Song Y, Timilsena PR, Van de Peer Y, Wang H, Wilhelmsson PKI, Wolf PG, Xu X, Der JP, Schluepmann H, Wong GK, Pryer KM ( 2018) Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nature Plants, 4, 460-472. |
| 25 | Lynch M, Conery JS ( 2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151-1155. |
| 26 | Muñoz A, Castellano MM ( 2012) Regulation of translation initiation under abiotic stress conditions in plants: It is a conserved or not so conserved process among eukaryotes? Comparative and Functional Genomics, 2012,406357. |
| 27 | Niu QK, Liang Y, Zhou JJ, Dou XY, Gao SC, Chen LQ, Zhang XQ, Ye D ( 2013) Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in Arabidopsis. Molecular Plant, 6, 1091-1098. |
| 28 | One Thousand Plant Transcriptomes Initiative ( 2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574, 679-685. |
| 29 | PPG I ( 2016) A community-derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution, 54, 563-603. |
| 30 | Rothfels CJ, Li FW, Sigel EM, Huiet L, Larsson A, Burge DO, Ruhsam M, Deyholos M, Soltis DE, Stewart CN Jr, Shaw SW, Pokorny L, Chen T, dePamphilis C, DeGironimo L, Chen L, Wei X, Sun X, Korall P, Stevenson DW, Graham SW, Wong GK, Pryer KM ( 2015) The evolutionary history of ferns inferred from 25 low-copy nuclear genes. American Journal of Botany, 102, 1089-1107. |
| 31 | Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC ( 2004) Mining EST databases to resolve evolutionary events in major crop species. Genome, 47, 868-876. |
| 32 | Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallon S, Lupia R ( 2004) Ferns diversified in the shadow of angiosperms. Nature, 428, 553-557. |
| 33 | Semon M, Wolfe KH ( 2007) Reciprocal gene loss between tetraodon and zebrafish after whole genome duplication in their ancestor. Trends in Genetics, 23, 108-112. |
| 34 | Smith AR, Cranfill RB ( 2002) Intrafamilial relationships of the thelypteroid ferns (Thelypteridaceae). American Fern Journal, 92, 131-149. |
| 35 | Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, depamphilis CW, Wall PK, Soltis PS ( 2009) Polyploidy and angiosperm diversification. American Journal of Botany, 96, 336-348. |
| 36 | Soltis PS, Marchant DB, Van de Peer Y, Soltis DE ( 2015) Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development, 35, 119-125. |
| 37 | Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z ( 2017) agriGO v2.0: A GO analysis toolkit for the agricultural community. Nucleic Acids Research, 45, 122-129. |
| 38 | Tsutsumi C, Kato M ( 2005) Molecular phylogenetic study on Davalliaceae. Fern Gazatte, 17, 147-162. |
| 39 | Tsutsumi C, Kato M ( 2006) Evolution of epiphytes in Davalliaceae and related ferns. Botanical Journal of the Linnean Society, 151, 495-510. |
| 40 | Vajda V, Raine JI, Hollis CJ ( 2001) Indication of global deforestation at the Cretaceous-Tertiary boundary by New Zealand fern spike. Science, 294, 1700-1702. |
| 41 | Van de Peer Y, Maere S, Meyer A ( 2009) The evolutionary significance of ancient genome duplications. Nature Reviews Genetics, 10, 725-732. |
| 42 | Vanneste K, Baele G, Maere S, Van de Peer Y ( 2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Research, 24, 1334-1347. |
| 43 | Vanneste K, Sterck L, Myburg AA, Van de Peer Y, Mizrachi E ( 2015) Horsetails are ancient polyploids: Evidence from Equisetum giganteum. The Plant Cell, 27, 1567-1578. |
| 44 | Vanneste K, Van de Peer Y, Maere S ( 2013) Inference of genome duplications from age distributions revisited. Molecular Biology and Evolution, 30, 177-190. |
| 45 | Wang JT, Li JT, Zhang XF, Sun XW ( 2012) Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genomics, 13, 96. |
| 46 | Wolfe KH, Shields DC ( 1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708-713. |
| 47 | Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH ( 2009) The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA, 106, 13875-13879. |
| 48 | Xiang YZ, Huang CH, Yi H, Wen J, Li S, Yi T, Chen H, Xiang J, Hong M ( 2017) Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Molecular Biology and Evolution, 34, 262-281. |
| 49 | Yang Z ( 2007) PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24, 1586-1591. |
| 50 | Zhang LB, Zhang L ( 2015) Didymochlaenaceae: A new fern family of eupolypods I (Polypodiales). Taxon, 64, 27-38. |
| 51 | Zhang R, Wang FG, Zhang J, Shang H, Liu L, Wang H, Zhao GH, Shen H, Yan YH ( 2019) Dating whole genome duplication in Ceratopteris thalictroides and potential adaptive values of retained gene duplicates. International Journal of Molecular Science, 20, 1926. |
| [1] | 陈楠, 张全国. 实验进化研究途径[J]. 生物多样性, 2024, 32(9): 24171-. |
| [2] | 葛颂. 中国植物系统和进化生物学研究进展[J]. 生物多样性, 2022, 30(7): 22385-. |
| [3] | 钱宏, 张健, 赵静超. 世界上已知维管植物有多少种? 基于多个全球植物数据库的整合[J]. 生物多样性, 2022, 30(7): 22254-. |
| [4] | 张淑梅, 李微, 李丁男. 辽宁省高等植物多样性编目[J]. 生物多样性, 2022, 30(6): 22038-. |
| [5] | 王婷, 夏增强, 舒江平, 张娇, 王美娜, 陈建兵, 王慷林, 向建英, 严岳鸿. 全基因组复制事件的绝对定年揭示莲座蕨属植物的迟滞演化[J]. 生物多样性, 2021, 29(6): 722-734. |
| [6] | 孙晶琦, 陈泉, 李航宇, 常艳芬, 巩合德, 宋亮, 卢华正. 附生蕨类植物的克隆性研究进展[J]. 生物多样性, 2019, 27(11): 1184-1195. |
| [7] | 詹臻, 张剑锋, 曹建国, 戴锡玲. 普通针毛蕨颈卵器和卵的发育[J]. 生物多样性, 2019, 27(11): 1236-1244. |
| [8] | 张开梅, 沈羽, 周晓丽, 方炎明. 21世纪以来蕨类植物研究论文的发表情况: 基于Web of Science的数据统计[J]. 生物多样性, 2019, 27(11): 1245-1250. |
| [9] | 姬红利, 詹选怀, 张丽, 彭焱松, 周赛霞, 胡菀. 幕阜山脉石松类和蕨类植物多样性及生物地理学特征[J]. 生物多样性, 2019, 27(11): 1251-1259. |
| [10] | 杜维波, 卢元. 黄土高原石松类和蕨类植物的多样性与地理分布[J]. 生物多样性, 2019, 27(11): 1260-1267. |
| [11] | 董仕勇, 左政裕, 严岳鸿, 向建英. 中国石松类和蕨类植物的红色名录评估[J]. 生物多样性, 2017, 25(7): 765-773. |
| [12] | 常艳芬. 铁角蕨科的多倍化与物种多样性形成[J]. 生物多样性, 2017, 25(6): 621-626. |
| [13] | 刘莉, 舒江平, 韦宏金, 张锐, 沈慧, 严岳鸿. 东亚特有珍稀蕨类植物岩穴蕨(碗蕨科)高通量转录组测序及分析[J]. 生物多样性, 2016, 24(12): 1325-1334. |
| [14] | 周喜乐, 张宪春, 孙久琼, 严岳鸿. 中国石松类和蕨类植物的多样性与地理分布[J]. 生物多样性, 2016, 24(1): 102-107. |
| [15] | 马克平. 生物多样性保护主流化的新机遇[J]. 生物多样性, 2015, 23(5): 557-558. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()