



生物多样性 ›› 2013, Vol. 21 ›› Issue (5): 616-627. DOI: 10.3724/SP.J.1003.2013.12059 cstr: 32101.14.SP.J.1003.2013.12059
所属专题: 物种形成与系统进化
收稿日期:2013-03-07
接受日期:2013-09-04
出版日期:2013-09-20
发布日期:2013-10-08
通讯作者:
裴男才
基金资助:Received:2013-03-07
Accepted:2013-09-04
Online:2013-09-20
Published:2013-10-08
Contact:
Pei Nancai
摘要:
DNA条形码为现代生物学研究提供了丰富的分子信息、标准的数据平台和通用的技术规程。本文从动物、植物和微生物等三方面简要梳理了生物DNA条形码近十年来的兴起和发展历程。参照DNA条形码特征, 将其生物学功能归纳为基本功能(如储备数据、鉴别物种)、延伸功能(如构建系统发育关系、服务特定行业、编制新一代生物图志)以及潜在功能(如物种整合)等三类。根据研究尺度, 划分出类群(主要是专科专属类研究)、群落(以自然保护区和大型固定样地构成的生物群落)和区域(生物多样性热点地区)等三个水平。列出了国际生命条形码组织开展的十大类研究项目, 从系统与分类学、生物多样性保护、系统发育进化生态学和数字化平台建设等四个方面分析了DNA条形码方法涉及的若干重要科学议题, 并指出在各学科应用时可能遇到的问题。DNA条形码技术在生物科学领域潜力巨大, 但还需在研究和论证过程中不断完善。
裴男才, 陈步峰 (2013) 生物DNA条形码: 十年发展历程、研究尺度和功能. 生物多样性, 21, 616-627. DOI: 10.3724/SP.J.1003.2013.12059.
Nancai Pei,Bufeng Chen (2013) DNA barcoding of life: a classification of uses according to function and scale after ten years of development. Biodiversity Science, 21, 616-627. DOI: 10.3724/SP.J.1003.2013.12059.
| 类别 Item | 片段名称 Barcode | 引物 Primer sequence (5′- 3′) | 来源 Source | 序列长度 Size (bp) | 参考文献 References |
|---|---|---|---|---|---|
| 动物 Animal | COI | 正向: GGTCAACAAATCATAAAGATATTGG 反向: TAAACTTCAGGGTGACCAAAAAATCA | 线粒体基因编码区 Coding region in mitochondrial gene | ~650 | Hebert et al., 2003a |
| 植物 Plant | rbcL matK trnH-psbA ITS trnL-F rps4 | 正向: ATGTCACCACAAACAGAGACTAAAGC 反向: GTYAAATCAAGTCCACCYCG 正向: CGTACAGTACTTTTGTGTTTACGAG 反向: ACCCAGTCCATCTGGAAATCTTGGTTC matK472F: CCCRTYCATCTGGAAATCTTGGTTC matK1248R: GCTRTRATAATGAGAAAGATTTCTGC trnH端: CGCGCATGGTGGATTCACAATCC psbA端: GTTATGCATGAACGTAATGCTC 正向: AGAAGTCGTAACAAGGTTTCCGTAGG 反向: TCCTCCGCTTATTGATATGC 正向: AGAAGTCGTAACAAGGTTTCCGTAGG或GGAAGGAGAAGTCGTAACAAGG; (裸子植物正向: GTCCACTGAACCTTATCATTTAG) 反向: TCCTCCGCTTATTGATATGC GTAAAACGACGGCCAGT CA-GGAA-ACAGCTATGAC | 叶绿体基因编码区 Coding region in chloroplast gene 叶绿体基因编码区 Coding region in chloroplast gene 叶绿体非编码基因间隔区 Non-coding intergenic region in chloroplast gene 核基因非编码基因间隔区 Non-coding intergenic region in nuclear gene 叶绿体非编码基因间隔区 Non-coding intergenic region in chloroplast gene 叶绿体基因编码区 Coding region in chloroplast gene | ~550 ~850 ~776 280-750 ~500(由多个较短序列组成) 350-500 700-900 | CBOL Plant Working Group, 2009; Kress & Erickson, 2007 Kim KJ, unpublished primers (http://www.korea.edu/academics/01co_04.php) Yu et al., 2011 Kress & Erickson, 2007 China Plant BOL Group, 2011 de Groot et al., 2011 Nadot et al., 1994 刘艳等, 2011 |
| 微生物 Microbe | ITS 18S rDNA | 正向: TCCTCCGCTTATTGATATGC 反向: GGAAGTAAAAGTCGTAACAAGG 正向: AGATTAAGCCATGCATGTCT 反向: GATCCTTCCGCAGGTTCACCTAC | 核基因非编码基因间隔区 Non-coding intergenic region in nuclear gene 核基因编码区 Coding region in nuclear gene | 400-660 1,600 | Seifert, 2009; Schoch et al., 2012; 张宇和郭良栋, 2012 Pawlowski et al., 2012 |
表1 常用生物DNA条形码片段一览表
Table 1 A list of currently used DNA barcodes of life
| 类别 Item | 片段名称 Barcode | 引物 Primer sequence (5′- 3′) | 来源 Source | 序列长度 Size (bp) | 参考文献 References |
|---|---|---|---|---|---|
| 动物 Animal | COI | 正向: GGTCAACAAATCATAAAGATATTGG 反向: TAAACTTCAGGGTGACCAAAAAATCA | 线粒体基因编码区 Coding region in mitochondrial gene | ~650 | Hebert et al., 2003a |
| 植物 Plant | rbcL matK trnH-psbA ITS trnL-F rps4 | 正向: ATGTCACCACAAACAGAGACTAAAGC 反向: GTYAAATCAAGTCCACCYCG 正向: CGTACAGTACTTTTGTGTTTACGAG 反向: ACCCAGTCCATCTGGAAATCTTGGTTC matK472F: CCCRTYCATCTGGAAATCTTGGTTC matK1248R: GCTRTRATAATGAGAAAGATTTCTGC trnH端: CGCGCATGGTGGATTCACAATCC psbA端: GTTATGCATGAACGTAATGCTC 正向: AGAAGTCGTAACAAGGTTTCCGTAGG 反向: TCCTCCGCTTATTGATATGC 正向: AGAAGTCGTAACAAGGTTTCCGTAGG或GGAAGGAGAAGTCGTAACAAGG; (裸子植物正向: GTCCACTGAACCTTATCATTTAG) 反向: TCCTCCGCTTATTGATATGC GTAAAACGACGGCCAGT CA-GGAA-ACAGCTATGAC | 叶绿体基因编码区 Coding region in chloroplast gene 叶绿体基因编码区 Coding region in chloroplast gene 叶绿体非编码基因间隔区 Non-coding intergenic region in chloroplast gene 核基因非编码基因间隔区 Non-coding intergenic region in nuclear gene 叶绿体非编码基因间隔区 Non-coding intergenic region in chloroplast gene 叶绿体基因编码区 Coding region in chloroplast gene | ~550 ~850 ~776 280-750 ~500(由多个较短序列组成) 350-500 700-900 | CBOL Plant Working Group, 2009; Kress & Erickson, 2007 Kim KJ, unpublished primers (http://www.korea.edu/academics/01co_04.php) Yu et al., 2011 Kress & Erickson, 2007 China Plant BOL Group, 2011 de Groot et al., 2011 Nadot et al., 1994 刘艳等, 2011 |
| 微生物 Microbe | ITS 18S rDNA | 正向: TCCTCCGCTTATTGATATGC 反向: GGAAGTAAAAGTCGTAACAAGG 正向: AGATTAAGCCATGCATGTCT 反向: GATCCTTCCGCAGGTTCACCTAC | 核基因非编码基因间隔区 Non-coding intergenic region in nuclear gene 核基因编码区 Coding region in nuclear gene | 400-660 1,600 | Seifert, 2009; Schoch et al., 2012; 张宇和郭良栋, 2012 Pawlowski et al., 2012 |
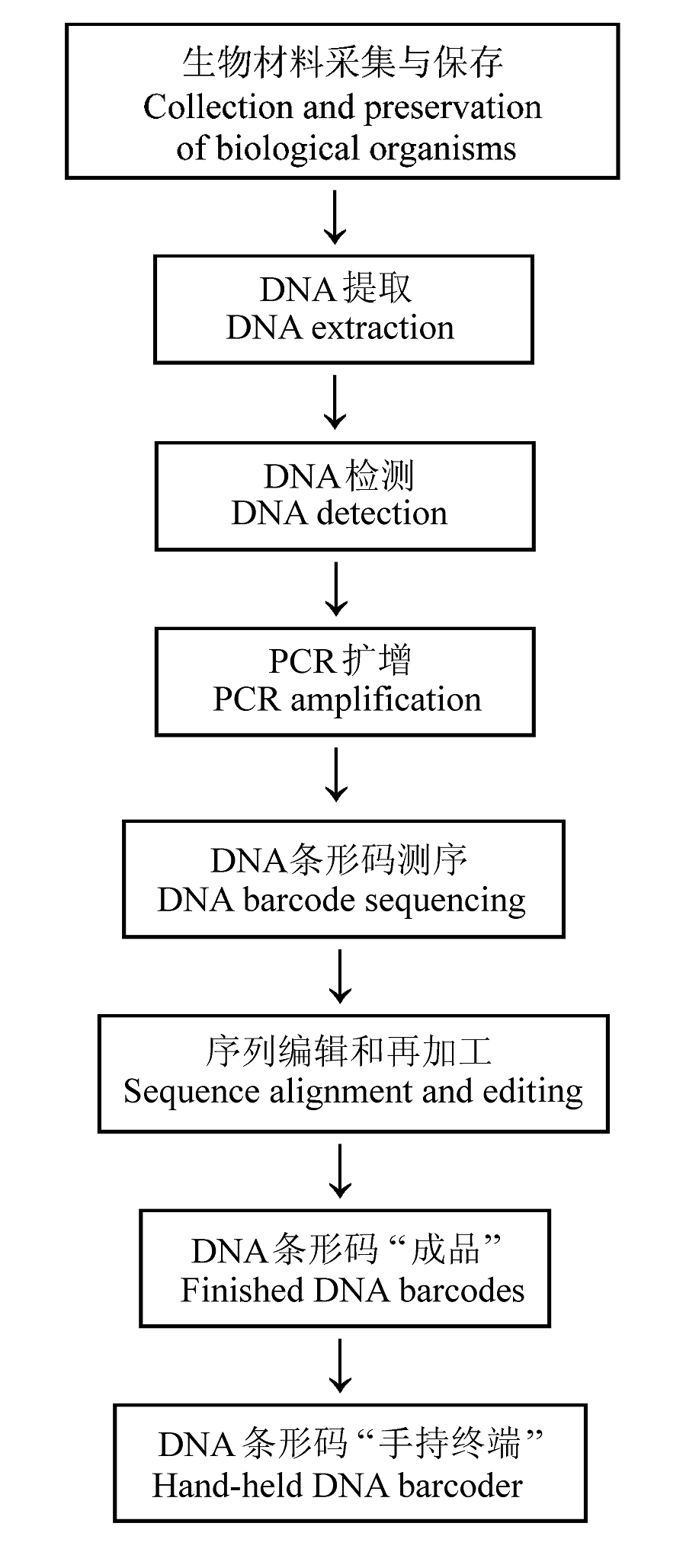
图1 DNA条形码的技术流程示意图(改自Janzen et al., 2009以及Paul Hebert教授于2009年在北京CBOL会议上使用的PPT)
Fig. 1 A graphic representation of DNA barcoding technological process (adopted from Janzen et al., 2009 and Prof. Paul Hebert’s PPT slides from CBOL Meeting in Beijing, May 2009)
| 1 | Albu M, Nikbakht H, Hajibabaei M, Hickey DA (2011) The DNA barcode linker.Molecular Ecology Resources, 11, 84-88. |
| 2 | Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Nucleic Acids Research, 25, 3389-3402. |
| 3 | Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, Hendrich L, Geijer J, Herrmann J, Foster GN, Ribera I, Nilsson AN, Barraclough TG, Vogler AP (2012) The effect of geographical scale of sampling on DNA barcoding.Systematic Biology, 61, 851-869. |
| 4 | Bhargava M, Sharma A (2013) DNA barcoding in plants: evolution and applications of in silico approaches and resources.Molecular Phylogenetics and Evolution, 67, 631-641. |
| 5 | Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation.Trends in Ecology and Evolution, 22, 148-155. |
| 6 | Bruni I, De Mattia F, Martellos S, Galimberti A, Savadori P, Casiraghi M, Nimis PL, Labra M (2012) DNA barcoding as an effective tool in improving a digital plant identification system: a case study for the area of Mt. Valerio, Trieste (NE Italy).PLoS ONE, 7, e43256. |
| 7 | Bucklin A, Steinke D, Blanco-Bercial L (2011) DNA barcoding of marine metazoa.Annual Review of Marine Science, 3, 471-508. |
| 8 | Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN (2008) DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservación Guanacaste, Costa Rica.Proceedings of the National Academy of Sciences, USA, 105, 6350-6355. |
| 9 | Cai YS (蔡延森), Zhang XY (张修月), Yue BS (岳碧松), Meng Y (孟杨), He LW (何莉炜), Li J (李静) (2009) DNA barcoding on the identification of 8 species of raptors.Sichuan Journal of Zoology(四川动物), 28, 334-340. (in Chinese with English abstract) |
| 10 | CBOL Plant Working Group (2009) A DNA barcode for land plants.Proceedings of the National Academy of Sciences , USA, 106, 12794-12797. |
| 11 | Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Savolainen V (2005) Land plants and DNA barcodes: short-term and long-term goals.Philosophical Transactions of the Royal Society B: Biological Sciences, 360, 1889-1895. |
| 12 | Che J, Chen HM, Yang JX, Jin JQ, Jiang K, Yuan ZY, Murphy RW, Zhang YP (2012) Universal COI primers for DNA barcoding amphibians.Molecular Ecology Resources, 12, 247-258. |
| 13 | Che J, Huang D, Li DZ, Ma JC, Zhang YP (2010) DNA barcoding and the international barcode of life project in China.Bulletin of the Chinese Academy of Sciences, 24, 257-260. |
| 14 | Chen J, Li Q, Kong LF, Yu H (2011) How DNA barcodes complement taxonomy and explore species diversity: the case study of a poorly understood marine fauna.PLoS ONE, 6, e21326. |
| 15 | Chen N (陈念), Zhao SJ (赵树进), Han LP (韩丽萍) (2008) Identification of fungi using DNA barcoding technology.International Journal of Laboratory Medicine(国际检验医学杂志), 29, 703-707. (in Chinese) |
| 16 | Chen SL (陈士林), Pang XH (庞晓慧), Luo K (罗焜), Yao H (姚辉), Han JP (韩建萍), Song JY (宋经元) (2013) DNA barcoding of biological resources.Chinese Bulletin of Life Sciences(生命科学), 25, 458-466. (in Chinese with English abstract) |
| 17 | Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species.PLoS ONE, 5, e8613. |
| 18 | Chen SL (陈士林), Yao H (姚辉), Song JY (宋经元), Li XW (李西文), Liu C (刘昶), Lu JW (陆建伟) (2007) Use of DNA barcoding to identify Chinese medicinal materials. World Science and Technology:Modernization of Traditional Chinese Medicine and Materia Medica(世界科学技术: 中医药现代化), 9(3), 7-12. (in Chinese with English abstract) |
| 19 | China Plant BOL Group (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants.Proceedings of the National Academy of Sciences, USA, 108, 19641-19646. |
| 20 | Collins RA, Cruickshank RH (2012) The seven deadly sins of DNA barcoding.Molecular Ecology Resources. doi: 10.1111/1755-0998.12046. |
| 21 | Craft KJ, Pauls SU, Darrow K, Miller SE, Hebert PDN, Helgen LE, Novotny V, Weiblen GD (2010) Population genetics of ecological communities with DNA barcodes: an example from New Guinea Lepidoptera.Proceedings of the National Academy of Sciences, USA, 107, 5041-5046. |
| 22 | Decaëns T, Porco D, Rougerie R, Brown GG, James SW (2013) Potential of DNA barcoding for earthworm research in taxonomy and ecology.Applied Soil Ecology, 65, 35-42. |
| 23 | de Groot GA, During HJ, Maas JW, Schneider H, Vogel JC, Erkens RHJ (2011) Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective.PLoS ONE, 6, e16371. |
| 24 | Dick CW, Kress WJ (2009) Dissecting tropical plant diversity with forest plots and a molecular toolkit.BioScience, 59, 745-755. |
| 25 | Elias M, Hill RI, Willmott KR, Dasmahapatra KK, Brower AVZ, Malllet J, Jiggins CD (2007) Limited performance of DNA barcoding in a diverse community of tropical butterflies.Proceedings of the Royal Society B: Biological Sciences, 274, 2881-2889. |
| 26 | Feng G, Zhang JL, Pei NC, Rao MD, Mi XC, Ren HB, Ma KP (2012) Comparison of phylobetadiversity indices based on community data from Gutianshan forest plot.Chinese Science Bulletin, 57, 623-630. |
| 27 | Galimberti A, De Mattia F, Losa A, Bruni I, Federici S, Casiraghi M, Martellos S, Labra M (2013) DNA barcoding as a new tool for food traceability.Food Research International, 50, 55-63. |
| 28 | Gao LM (高连明), Liu J (刘杰), Cai J (蔡杰), Yang JB (杨俊波), Zhang T (张挺), Li DZ (李德铢) (2012) A synopsis of technical notes on the standards for plant DNA barcoding.Plant Diversity and Resources(植物分类与资源学报), 34, 592-606. (in Chinese with English abstract) |
| 29 | Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA (2007) The current status of species recognition and identification in Aspergillus.Studies in Mycology, 59, 1-10. |
| 30 | Ghahramanzadeh R, Esselink G, Kodde LP, Duistermaat H, van Valkenburg JLCH, Marashi SH, Smulders MJM, van de Wiel CCM (2013) Efficient distinction of invasive aquatic plant species from non-invasive related species using DNA barcoding.Molecular Ecology Resources, 13, 21-31. |
| 31 | Gilmore SR, Gräfenhan T, Louis-Seize G, Seifert KA (2009) Multiple copies of cytochrome oxidase 1 in species of the fungal genus Fusarium.Molecular Ecology Resources, 9, 90-98. |
| 32 | Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Riéra B, Roger A, Thébaud C, Chave J (2009) Identification of Amazonian trees with DNA barcodes.PLoS ONE, 4, e7483. |
| 33 | Grant RA, Griffiths HJ, Steinke D, Wadley V, Linse K (2011) Antarctic DNA barcoding: a drop in the ocean? Polar Biology, 34, 775-780. |
| 34 | Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera.Proceedings of the National Academy of Sciences, USA, 103, 968-971. |
| 35 | He K (何锴), Wang WZ (王文智), Li Q (李权), Luo PP (罗培鹏), Sun YH (孙悦华), Jiang XL (蒋学龙) (2013) DNA barcoding in surveys of small mammal community: a case study in Lianhuashan, Gansu Province, China.Biodiversity Science(生物多样性), 21, 197-205. (in Chinese with English abstract) |
| 36 | Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003a) Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. |
| 37 | Hebert PDN, Gregory TR (2005) The promise of DNA barcoding for taxonomy.Systematic Biology, 54, 852-859. |
| 38 | Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004a) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator.Proceedings of the National Academy of Sciences, USA, 101, 14812-14817. |
| 39 | Hebert PDN, Ratnasingham S, deWaard JR (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species.Proceedings of the Royal Society B: Biological Sciences, 270, S96-S99. |
| 40 | Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004b) Identification of birds through DNA barcodes.PLoS Biology, 2, 1657-1663. |
| 41 | Horton TR, Bruns TD (2001) The molecular revolution in ectomycorrhizal ecology: peeking into the black-box.Molecular Ecology, 10, 1855-1871. |
| 42 | Hsu TH, Ning Y, Gwo JC, Zeng ZN (2013) DNA barcoding reveals cryptic diversity in the peanut worm Sipunculus nudus.Molecular Ecology Resources, 13, 596-606. |
| 43 | Janzen DH, Hallwachs W, Blandin P, Burns JM, Cadiou JM, Chacon I, Dapkey T, Deans AR, Epstein ME, Espinoza B, Franclemont JG, Haber WA, Hajibabaei M, Hall JPW, Hebert PDN, Gauld ID, Harvey DJ, Hausmann A, Kitching IJ, Lafontaine D, Landry JF, Lemaire C, Miller JY, Miller JS, Miller L, Miller SE, Montero J, Munroe E, Green SR, Ratnasingham S, Rawlins JE, Robbins RK, Rodriguez JJ, Rougerie R, Sharkey MJ, Smith MA, Solis MA, Sullivan JB, Thiaucourt P, Wahl DB, Weller SJ, Whitfield JB, Willmott KR, Wood DM, Woodley NE, Wilson JJ (2009) Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity.Molecular Ecology Resources, 9, 1-26. |
| 44 | Jögrer K, Norenburg J, Wilson N, Schrödl M (2012) Barcoding against a paradox? Combined molecular species delineations reveal multiple cryptic lineages in elusive meiofaunal sea slugs.BMC Evolutionary Biology, 12, 245. |
| 45 | Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region.PLoS ONE, 2, e508. |
| 46 | Kress WJ, Erickson DL (2012) DNA Barcodes: Methods and Protocols. Humana Press, Berlin. |
| 47 | Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E (2009) Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama.Proceedings of the National Academy of Sciences , USA, 106, 18621-18626. |
| 48 | Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants.Proceedings of the National Academy of Sciences, USA, 102, 8369-8374. |
| 49 | Kwong S, Srivathsan A, Meier R (2012) An update on DNA barcoding: low species coverage and numerous unidentified sequences.Cladistics, 28, 639-644. |
| 50 | Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots.Proceedings of the National Academy of Sciences, USA, 105, 2923-2928. |
| 51 | Li CL (李超伦), Wang MX (王敏晓), Cheng FP (程方平), Sun S (孙松) (2011) DNA barcoding and its application to marine zooplankton ecology.Biodiversity Science(生物多样性) 19, 805-814. (in Chinese with English abstract) |
| 52 | Li DZ, Liu JQ, Chen ZD, Wang H, Ge XJ, Zhou SL, Gao LM, Fu CX, Chen SL (2011) Plant DNA barcoding in China.Journal of Systematics and Evolution, 49, 165-168. |
| 53 | Li DZ (李德铢), Wang YH (王雨华), Yi TS (伊廷双), Wang H (王红), Gao LM (高连明), Yang JB (杨俊波) (2012) The next generation flora: iFlora.Plant Diversity and Resources(植物分类与资源学报), 34, 525-531. (in Chinese with English abstract) |
| 54 | Lijtmaer DA, Kerr KCR, Barreira AS, Hebert PDN, Tubaro PL (2011) DNA barcode libraries provide insight into continental patterns of avian diversification.PLoS ONE, 6, e20744. |
| 55 | Liu Y (刘艳), Wang JX (王建秀), Ge XJ (葛学军), Cao T (曹同) (2011) The rps4 locus as an alternative marker for barcoding bryophytes: evaluation based on data mining from GenBank.Biodiversity Science(生物多样性), 19, 311-318. (in Chinese with English abstract) |
| 56 | Ma MY (马明义), Yan Y (闫颖), Wang YW (王译伟), Li J (李静), Cai YS (蔡延森), Li JL (李佳凌) (2012) A study of DNA barcoding on 32 species of bird in China.Sichuan Journal of Zoology(四川动物), 31, 729-733. (in Chinese with English abstract) |
| 57 | Mayfield MM, Boni MF, Ackerly DD (2009) Traits, habitats, and clades: identifying traits of potential importance to environmental filtering.The American Naturalist, 174, E1-E22. |
| 58 | Meier R, Shiyang K, Vaidya G, Ng PKL (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success.Systematic Biology, 55, 715-728. |
| 59 | Moritz C, Cicero C (2004) DNA barcoding: promise and pitfalls.PLoS Biology, 2, e354. |
| 60 | Murphy RW, Crawford AJ, Bauer AM, Che J, Donnellan SC, Fritz U, Haddad CFB, Nagy ZT, Poyarkov NA, Vences M, Wang WZ, Zhang YP (2013) Cold Code: the global initiative to DNA barcode amphibians and nonavian reptiles.Molecular Ecology Resources, 13, 161-167. |
| 61 | Myers N, Mittermeier RA, Mittermeier CG, Fonseca GABd, Kent J (2000) Biodiversity hotspots for conservation priorities.Nature, 403, 853-858. |
| 62 | Nadot S, Bajon R, Lejeune B (1994) The chloroplast gene rps4 as a tool for the study of Poaceae phylogeny.Plant Systematics and Evolution, 191, 27-38. |
| 63 | Ning SP (宁淑萍), Yan HF (颜海飞), Hao G (郝刚), Ge XJ (葛学军) (2008) Current advances of DNA barcoding study in plants.Biodiversity Science(生物多样性), 16, 417-425. (in Chinese with English abstract) |
| 64 | Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Cepicka I, Decelle J, Dunthorn M, Fiore-Donno AM, Gile GH, Holzmann M, Jahn R, Jirků M, Keeling PJ, Kostka M, Kudryavtsev A, Lara E, Lukeš J, Mann DG, Mitchell EAD, Nitsche F, Romeralo M, Saunders GW, Simpson AGB, Smirnov AV, Spouge JL, Stern RF, Stoeck T, Zimmermann J, Schindel D, de Vargas C (2012) CBOL Protist Working Group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms.PLoS Biology, 10, e1001419. |
| 65 | Pei NC (裴男才) (2012a) Identification of plant species based on DNA barcode technology.Chinese Journal of Applied Ecology(应用生态学报), 23, 1240-1246. (in Chinese with English abstract) |
| 66 | Pei NC (裴男才) (2012b) Building a subtropical forest community phylogeny based on plant DNA barcodes from Dinghushan plot.Plant Diversity and Resources(植物分类与资源学报), 34, 263-270. (in Chinese with English abstract) |
| 67 | Pei NC, Lian JY, Erickson DL, Swenson NG, Kress WJ, Ye WH, Ge XJ (2011) Exploring tree-habitat associations in a Chinese subtropical forest plot using a molecular phylogeny generated from DNA barcode loci.PLoS ONE, 6, e21273. |
| 68 | Pei NC (裴男才), Zhang JL (张金龙), Mi XC (米湘成), Ge XJ (葛学军) (2011) Plant DNA barcodes promote the development of phylogenetic community ecology.Biodiversity Science(生物多样性), 19, 284-294. (in Chinese with English abstract) |
| 69 | Ren BQ (任保青), Chen ZD (陈之端) (2010) DNA barcoding plant life.Chinese Bulletin of Botany(植物学报), 45, 1-12. (in Chinese with English abstract) |
| 70 | Sarkar IN, Trizna M (2011) The barcode of life data portal: bridging the biodiversity informatics divide for DNA barcoding.PLoS ONE, 6, e14689. |
| 71 | Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Consortium FB (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.Proceedings of the National Academy of Sciences, USA, 109, 6241-6246. |
| 72 | Seifert KA (2009) Progress towards DNA barcoding of fungi.Molecular Ecology Resources, 9, 83-89. |
| 73 | Shen YY, Chen X, Murphy RW (2013) Assessing DNA barcoding as a tool for species identification and data quality control.PLoS ONE, 8, e57125. |
| 74 | Smith MA, Fernandez-Triana J, Roughley R, Hebert PDN (2009) DNA barcode accumulation curves for understudied taxa and areas.Molecular Ecology Resources, 9, 208-216. |
| 75 | Swenson NG, Erickson DL, Mi XC, Bourg NA, Forero-Montaña J, Ge XJ, Howe R, Lake JK, Liu XJ, Ma KP, Pei NC, Thompson J, Uriarte M, Wolf A, Wright SJ, Ye WH, Zhang JL, Zimmerman JK, Kress WJ (2012) Phylogenetic and functional alpha and beta diversity in temperate and tropical tree communities.Ecology, 93, S112-S125. |
| 76 | Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding.Nucleic Acids Research, 35, e14. |
| 77 | van de Wiel CCM, van der Schoot J, van Valkenburg JLCH, Duistermaat H, Smulders MJM (2009) DNA barcoding discriminates the noxious invasive plant species, floating pennywort (Hydrocotyle ranunculoides L. f.), from non-invasive relatives.Molecular Ecology Resources, 9, 1086-1091. |
| 78 | von Cräutlein M, Korpelainen H, Pietiläinen M, Rikkinen J (2011) DNA barcoding: a tool for improved taxon identification and detection of species diversity.Biodiversity and Conservation, 20, 373-389. |
| 79 | Wang G, Li CX, Guo X, Xing D, Dong YD, Wang ZM, Zhang YM, Liu MD, Zheng Z, Zhang HD, Zhu XJ, Wu ZM, Zhao TY (2012) Identifying the main mosquito species in China based on DNA barcoding.PLoS ONE, 7, e47051. |
| 80 | Wang JF (王剑峰), Qiao GX (乔格侠) (2007) Application of DNA barcoding to Aphidinea (Insecta, Hemiptera) studies.Acta Zootaxonomica Sinica(动物分类学报), 32, 153-159. (in Chinese with English abstract) |
| 81 | Wang Q, Guo LD (2010) Ectomycorrhizal community composition of Pinus tabulaeformis assessed by ITS-RFLP and ITS sequences.Botany, 88, 590-595. |
| 82 | Wang WJ (王文婧), Wang XL (王晓亮), Wang XC (王新存), Yu XD (于晓丹), Li Y (李熠), Wei TZ (魏铁铮), Yao YJ (姚一建) (2009) Application of DNA barcoding in fungal research.Frontier Science(前沿科学), 3(2), 4-12. (in Chinese with English abstract) |
| 83 | Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species.Proceedings of the Royal Society B: Biological Sciences, 360, 1847-1857. |
| 84 | Waugh J (2007) DNA barcoding in animal species: progress, potential and pitfalls.BioEssays, 29, 188-197. |
| 85 | Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology.Annual Review of Ecology and Systematics, 33, 475-505. |
| 86 | Will KW, Mishler BD, Wheeler QD (2005) The perils of DNA barcoding and the need for integrative taxonomy.Systematic Biology, 54, 844-851. |
| 87 | Xiao JH (肖金花), Xiao H (肖晖), Huang DW (黄大卫) (2004) DNA barcoding: new approach of biological taxonomy.Acta Zoologica Sinica(动物学报), 50, 852-855. (in Chinese with English abstract) |
| 88 | Yu J, Xue J-H, Zhou S-L (2011) New universal matK primers for DNA barcoding angiosperms.Journal of Systematics and Evolution, 49, 176-181. |
| 89 | Zhang JM (张金梅), Wang JX (王建秀), Xia T (夏涛), Zhou SL (周世良) (2008) Application of DNA barcoding technology in clarificating the species issues in Paeonia sect. Moutan based on phylogenetic analyses.Scientia Sinica Vitae(中国科学, 38, 1166-1176. (in Chinese) |
| 90 | Zhang Y (张宇), Guo LD (郭良栋) (2012) Progress of fungal DNA barcode.Mycosystema(菌物学报), 31, 809-820. (in Chinese with English abstract) |
| 91 | Zhuang HF (庄会富), Wang YH (王雨华) (2012) A framework of scientific-cloud for iFlora.Plant Diversity and Resources(植物分类与资源学报), 34, 623-630. (in Chinese with English abstract) |
| [1] | 祝晓雨, 王晨灏, 王忠君, 张玉钧. 城市绿地生物多样性研究进展与展望[J]. 生物多样性, 2025, 33(5): 25027-. |
| [2] | 王欣, 鲍风宇. 基于鸟类多样性提升的南滇池国家湿地公园生态修复效果分析[J]. 生物多样性, 2025, 33(5): 24531-. |
| [3] | 明玥, 郝培尧, 谭铃千, 郑曦. 基于城市绿色高质量发展理念的中国城市生物多样性保护与提升研究[J]. 生物多样性, 2025, 33(5): 24524-. |
| [4] | 卢晓强, 董姗姗, 马月, 徐徐, 邱凤, 臧明月, 万雅琼, 李孪鑫, 于赐刚, 刘燕. 前沿技术在生物多样性研究中的应用现状、挑战与展望[J]. 生物多样性, 2025, 33(4): 24440-. |
| [5] | 赵维洋, 王伟, 马冰然. 其他有效的区域保护措施(OECMs)研究进展与展望[J]. 生物多样性, 2025, 33(3): 24525-. |
| [6] | 周志华, 金效华, 罗颖, 李迪强, 岳建兵, 刘芳, 何拓, 李希, 董晖, 罗鹏. 中国林草部门落实《昆明-蒙特利尔全球生物多样性框架》的机制、成效分析及建议[J]. 生物多样性, 2025, 33(3): 24487-. |
| [7] | 刘立, 臧明月, 马月, 万雅琼, 胡飞龙, 卢晓强, 刘燕. 央地协同推动国家生物多样性战略和行动计划执行的措施、进展与展望[J]. 生物多样性, 2025, 33(3): 24532-. |
| [8] | 宋阳, 柳军, 何少林, 徐薇, 程琛, 刘博, 余绩庆. 我国能源企业生物多样性保护主流化管理路径[J]. 生物多样性, 2025, 33(1): 24345-. |
| [9] | 苏荣菲, 陈睿山, 俞霖琳, 吴婧彬, 康燕. 基于红外相机调查的上海市长宁区社区生境花园生物多样性[J]. 生物多样性, 2024, 32(8): 24068-. |
| [10] | 蔡颖莉, 朱洪革, 李家欣. 中国生物多样性保护政策演进、主要措施与发展趋势[J]. 生物多样性, 2024, 32(5): 23386-. |
| [11] | 鄢德奎. 中国生物多样性保护政策的共同要素、不足和优化建议[J]. 生物多样性, 2024, 32(5): 23293-. |
| [12] | 刘荆州, 钱易鑫, 张燕雪丹, 崔凤. 基于潜在迪利克雷分布(LDA)模型的旗舰物种范式研究进展与启示[J]. 生物多样性, 2024, 32(4): 23439-. |
| [13] | 陈越, 毛子昆, 王绪高. 基于生态独特性的β多样性研究进展与未来展望[J]. 生物多样性, 2024, 32(12): 24199-. |
| [14] | 张梓欣, 张承云, 郝泽周, 何凯莹, 黄泳桥, 肖治术. 陆地生物声学数据采集设备的进展及展望[J]. 生物多样性, 2024, 32(10): 24265-. |
| [15] | 刘莹莹, 龚立新, 曾皓, 冯江, 董永军, 王磊, 江廷磊. 被动声学监测在蝙蝠研究中的应用[J]. 生物多样性, 2024, 32(10): 24233-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()
