



Biodiv Sci ›› 2019, Vol. 27 ›› Issue (8): 842-853. DOI: 10.17520/biods.2019034 cstr: 32101.14.biods.2019034
Previous Articles Next Articles
Zhongdong Yu1,Zhihe Yu2,Shiyu Jin3,Long Wang4,*( )
)
Received:2019-02-14
Accepted:2019-05-15
Online:2019-08-20
Published:2019-05-20
Contact:
Wang Long
Zhongdong Yu, Zhihe Yu, Shiyu Jin, Long Wang. Genetic diversity and toxin-producing characters of Aspergillus flavus from China[J]. Biodiv Sci, 2019, 27(8): 842-853.
| 菌株顺序号* Number | 物种 Species | 菌株 Strains | 分离地和基物 Isolation places and substrates | 产毒素 Toxin production | |
|---|---|---|---|---|---|
| AFB | CPA | ||||
| 1 | A. flavus | CBS 100927T | 南太平洋群岛; 赛璐玢 Cellophane; South Pacific Islands | - | + |
| 2 | A. oryzae | CBS 100925T | 日本大阪; 分离基物未知 Ex-type of A. oryzae, unknown source; Osaka, Japan | - | + |
| 3 | A. thomii | CBS 120.51T | 英国伦敦; 污染物 Ex-type of A. thomii, culture contaminant; London, UK | - | + |
| 4 | A. flavus | CYH2-2-1 | 河北石家庄; 空气 Air; Shijiazhuang, Hebei, China | + | + |
| 5 | A. flavus | 3.4408 | 日本东京; 土壤 Soil; Tokyo, Japan | + | + |
| 6 | A. flavus | 14527 | 西藏米林; 土壤 Soil; Milin, Tibet, China | + | + |
| 7 | A. flavus | 13483 | 山西五台山; 土壤 Soil; Mt. Wutaishan, Shanxi, China | + | + |
| 8 | A. flavus | 13868 | 内蒙古呼伦贝尔; 土壤 Soil; Hulun Buir, Inner Mongolia, China | + | + |
| 9 | A. flavus | 13894 | 湖南益阳; 土壤 Soil; Yiyang, Hunan, China | + | + |
| 10 | A. flavus | 13895 | 江西三清山; 土壤 Soil; Sanqingshan, Jiangxi, China | + | + |
| 11 | A. flavus | 13918 | 江苏苏州; 土壤 Soil, Suzhou, Jiangsu, China | + | + |
| 12 | A. flavus | 13952 | 甘肃兰州; 土壤 Soil; Lanzhou, Gansu, China | - | + |
| 13 | A. flavus | 13961 | 宁夏罗山; 土壤 Soil; Luoshan, Ningxia, China | - | - |
| 14 | A. flavus | 13962 | 宁夏灵武; 土壤 Soil; Lingwu, Ningxia, China | - | - |
| 15 | A. flavus | 14099 | 山西吕梁; 土壤 Soil; lvliang, Shanxi, China | + | + |
| 16 | A. flavus | 14131 | 山西大同; 土壤 Soil; Datong, Shanxi, China | + | + |
| 17 | A. flavus | 14151 | 河南洛阳; 土壤 Soil; Luoyang, Henan, China | - | - |
| 18 | A. flavus | 14152 | 河南南阳; 土壤 Soil; Nanyang, Henan, China | - | - |
| 19 | A. flavus | 14153 | 山东泰安 土壤 Soil; Tai’an, Shandong, China | - | - |
| 20 | A. flavus | 14154 | 山东临沂; 土壤 Soil; Linyi, Shandong, China | - | - |
| 21 | A. flavus | 14155 | 河北兴隆; 土壤 Soil; Xinglong, Hebei, China | - | - |
| 22 | A. flavus | 14156 | 河北张家口; 土壤 Soil; Zhangjiakou, Hebei, China | - | - |
| 23 | A. flavus | 14157 | 河北保定; 土壤 Soil; Baoding, Hebei, China | - | - |
| 24 | A. flavus | 14159 | 河北衡水; 土壤 Soil; Hengshui, Hebei, China | - | - |
| 25 | A. flavus | 14175 | 浙江乌镇; 土壤 Soil; Wuzhen, Zhejiang, China | + | + |
| 26 | A. flavus | 14334 | 安徽巢湖; 荸荠 Water chestnut; Chaohu, Anhui, China | + | + |
| 27 | A. flavus | 14353 | 新疆吐鲁番; 土壤 Soil; Turpan, Xinjiang, China | + | + |
| 28 | A. flavus | 14355 | 新疆石河子; 土壤 Soil; Shihezi, Xinjiang, China | + | + |
| 29 | A. flavus | 14356 | 新疆乌鲁木齐; 土壤 Soil; Urumqi, Xinjiang, China | + | + |
| 30 | A. flavus | 14357 | 新疆伊犁; 土壤 Soil; Yili, Xinjiang, China | + | + |
| 31 | A. flavus | 14358 | 陕西榆林; 土壤 Soil; Yulin, Shaanxi, China | + | + |
| 32 | A. flavus | 14359 | 陕西汉中; 土壤 Soil; Hanzhong, Shaanxi, China | + | + |
| 33 | A. flavus | 14373 | 陕西渭南; 土壤 Soil; Weinan, Shaanxi, China | + | + |
| 34 | A. flavus | 14374 | 新疆吐鲁番; 土壤 Soil, Turpan, Xinjiang, China | + | + |
| 35 | A. flavus | 23124 | 海南五指山; 土壤 Soil; Mt. Wuzhishan, Hainan, China | + | + |
Table 1 Aspergillus flavus strains, isolation places and toxin-production
| 菌株顺序号* Number | 物种 Species | 菌株 Strains | 分离地和基物 Isolation places and substrates | 产毒素 Toxin production | |
|---|---|---|---|---|---|
| AFB | CPA | ||||
| 1 | A. flavus | CBS 100927T | 南太平洋群岛; 赛璐玢 Cellophane; South Pacific Islands | - | + |
| 2 | A. oryzae | CBS 100925T | 日本大阪; 分离基物未知 Ex-type of A. oryzae, unknown source; Osaka, Japan | - | + |
| 3 | A. thomii | CBS 120.51T | 英国伦敦; 污染物 Ex-type of A. thomii, culture contaminant; London, UK | - | + |
| 4 | A. flavus | CYH2-2-1 | 河北石家庄; 空气 Air; Shijiazhuang, Hebei, China | + | + |
| 5 | A. flavus | 3.4408 | 日本东京; 土壤 Soil; Tokyo, Japan | + | + |
| 6 | A. flavus | 14527 | 西藏米林; 土壤 Soil; Milin, Tibet, China | + | + |
| 7 | A. flavus | 13483 | 山西五台山; 土壤 Soil; Mt. Wutaishan, Shanxi, China | + | + |
| 8 | A. flavus | 13868 | 内蒙古呼伦贝尔; 土壤 Soil; Hulun Buir, Inner Mongolia, China | + | + |
| 9 | A. flavus | 13894 | 湖南益阳; 土壤 Soil; Yiyang, Hunan, China | + | + |
| 10 | A. flavus | 13895 | 江西三清山; 土壤 Soil; Sanqingshan, Jiangxi, China | + | + |
| 11 | A. flavus | 13918 | 江苏苏州; 土壤 Soil, Suzhou, Jiangsu, China | + | + |
| 12 | A. flavus | 13952 | 甘肃兰州; 土壤 Soil; Lanzhou, Gansu, China | - | + |
| 13 | A. flavus | 13961 | 宁夏罗山; 土壤 Soil; Luoshan, Ningxia, China | - | - |
| 14 | A. flavus | 13962 | 宁夏灵武; 土壤 Soil; Lingwu, Ningxia, China | - | - |
| 15 | A. flavus | 14099 | 山西吕梁; 土壤 Soil; lvliang, Shanxi, China | + | + |
| 16 | A. flavus | 14131 | 山西大同; 土壤 Soil; Datong, Shanxi, China | + | + |
| 17 | A. flavus | 14151 | 河南洛阳; 土壤 Soil; Luoyang, Henan, China | - | - |
| 18 | A. flavus | 14152 | 河南南阳; 土壤 Soil; Nanyang, Henan, China | - | - |
| 19 | A. flavus | 14153 | 山东泰安 土壤 Soil; Tai’an, Shandong, China | - | - |
| 20 | A. flavus | 14154 | 山东临沂; 土壤 Soil; Linyi, Shandong, China | - | - |
| 21 | A. flavus | 14155 | 河北兴隆; 土壤 Soil; Xinglong, Hebei, China | - | - |
| 22 | A. flavus | 14156 | 河北张家口; 土壤 Soil; Zhangjiakou, Hebei, China | - | - |
| 23 | A. flavus | 14157 | 河北保定; 土壤 Soil; Baoding, Hebei, China | - | - |
| 24 | A. flavus | 14159 | 河北衡水; 土壤 Soil; Hengshui, Hebei, China | - | - |
| 25 | A. flavus | 14175 | 浙江乌镇; 土壤 Soil; Wuzhen, Zhejiang, China | + | + |
| 26 | A. flavus | 14334 | 安徽巢湖; 荸荠 Water chestnut; Chaohu, Anhui, China | + | + |
| 27 | A. flavus | 14353 | 新疆吐鲁番; 土壤 Soil; Turpan, Xinjiang, China | + | + |
| 28 | A. flavus | 14355 | 新疆石河子; 土壤 Soil; Shihezi, Xinjiang, China | + | + |
| 29 | A. flavus | 14356 | 新疆乌鲁木齐; 土壤 Soil; Urumqi, Xinjiang, China | + | + |
| 30 | A. flavus | 14357 | 新疆伊犁; 土壤 Soil; Yili, Xinjiang, China | + | + |
| 31 | A. flavus | 14358 | 陕西榆林; 土壤 Soil; Yulin, Shaanxi, China | + | + |
| 32 | A. flavus | 14359 | 陕西汉中; 土壤 Soil; Hanzhong, Shaanxi, China | + | + |
| 33 | A. flavus | 14373 | 陕西渭南; 土壤 Soil; Weinan, Shaanxi, China | + | + |
| 34 | A. flavus | 14374 | 新疆吐鲁番; 土壤 Soil, Turpan, Xinjiang, China | + | + |
| 35 | A. flavus | 23124 | 海南五指山; 土壤 Soil; Mt. Wuzhishan, Hainan, China | + | + |
| 菌株顺序号* Number | 物种 Species | 菌株 Strains | 分离地和基物 Isolation places and substrates | 产毒素 Toxin production | |
|---|---|---|---|---|---|
| AFB | CPA | ||||
| 36 | A. flavus | AB34 | 四川若尔盖; 土壤 Soil; Ruoergai Prairie, Sichuan, China | - | + |
| 37 | A. flavus | FJ17-2 | 福建宁德; 茶叶 Tea; Ningde, Fujian, China | + | + |
| 38 | A. flavus | HB4 | 湖北神农架; 土壤 Soil; Shennongjia, Hubei, China | + | + |
| 39 | A. flavus | HL53 | 黑龙江凉水; 土壤 Soil; Liangshui Nature Reserve, Heilongjiang, China | + | + |
| 40 | A. flavus | HL70 | 黑龙江乌伊岭; 土壤 Soil; Wuyiling, Heilongjiang, China | + | + |
| 41 | A. flavus | KK39 | 青海互助县; 土壤 Soil; Huzhu County, Qinghai, China | - | + |
| 42 | A. flavus | KK41 | 青海互助县; 土壤 Soil; Huzhu County, Qinghai, China | - | + |
| 43 | A. flavus | KK49 | 青海互助县; 土壤 Soil; Huzhu County, Qinghai, China | - | + |
| 44 | A. flavus | KK50 | 青海可可西里; 土壤 Soil; Hoh Xil, Qinghai, China | - | + |
| 45 | A. flavus | KK65 | 青海楚玛尔河; 土壤 Soil; Chumaer River, Qinghai, China | - | + |
| 46 | A. flavus | KK66 | 青海可可西里; 土壤 Soil; Hoh Xil, Qinghai, China | - | + |
| 47 | A. flavus | KK67 | 青海楚玛尔河; 土壤 Soil; Chumaer River, Qinghai, China | - | + |
| 48 | A. flavus | KK68 | 青海沱沱河; 土壤 Soil; Tuotuo River, Qinghai, China | - | + |
| 49 | A. flavus | KK69 | 青海沱沱河; 土壤 Soil; Tuotuo River, Qinghai, China | - | + |
| 50 | A. flavus | KK70 | 青海楚玛尔河; 土壤 Soil; Chumaer River, Qinghai, China | - | + |
| 51 | A. flavus | KK72 | 青海青海湖; 土壤 Soil; Qinghai Lake, Qinghai, China | + | + |
| 52 | A. flavus | KK73 | 青海青海湖; 土壤 Soil; Qinghai Lake, Qinghai, China | + | + |
| 53 | A. flavus | KK94 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | + | + |
| 54 | A. flavus | KK102 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 55 | A. flavus | KK103 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 56 | A. flavus | KK104 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 57 | A. flavus | KK114 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 58 | A. flavus | XZ107 | 陕西南宫山; 植物叶 Plant leaves; Mt. Nangongshan, Shaanxi, China | + | + |
| 59 | A. flavus | XZ108 | 陕西通天河; 植物叶 Plant leaves; Tongtian River, Shaanxi, China | + | + |
| 60 | A. flavus | XZ109 | 陕西南宫山; 植物叶 Plant leaves; Mt. Nangongshan, Shaanxi, China | + | + |
| 61 | A. flavus | XZ112 | 陕西通天河; 植物叶 Plant leaves; Tongtian River, Shaanxi, China | + | + |
| 62 | A. flavus | YN23 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | - | + |
| 63 | A. flavus | YN35 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 64 | A. flavus | YN48 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 65 | A. flavus | YN49 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 66 | A. flavus | YN51 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 67 | A. flavus | NRRL 3357 | 美国; 霉花生 Moldy peanuts; USA | + | + |
| 68 | A. oryzae | RIB40 | 日本; 谷粒 Cereal grains; Japan | - | - |
| 69 | A. flavus | 3.262 | 辽宁大连; 空气 Air; Dalian, Liaoning, China | - | - |
| 70 | A. flavus | 3.267 | 辽宁大连; 土壤 Soil; Dalian, Liaoning, China | - | + |
| 71 | A. flavus | 3.337 | 天津; 蚊香 Mosquito-repellent incense; Tianjin, China | - | + |
| 72 | A. flavus | 3.417 | 天津; 酱曲 Soy sauce starter; Tianjin, China | - | + |
| 73 | A. flavus | 3.870 | 天津; 酱曲 Soy sauce starter; Tianjin, China | + | + |
| 74 | A. flavus | 3.881 | 上海; 小麦 Wheat; Shanghai, China | - | + |
| 75 | A. flavus | 3.2146 | 北京; 大米 Rice; Beijing, China | + | + |
| 76 | A. flavus | 3.2758 | 广东广州; 空气 Air; Guangzhou, Guangdong, China | - | + |
| 77 | A. flavus | 3.2789 | 越南河内; 土壤 Soil; Hanoi, Vietnam | - | + |
| 78 | A. flavus | 3.2823 | 安徽芜湖; 植物 Plants; Wuhu, Anhui, China | - | + |
Table 1 (continued)
| 菌株顺序号* Number | 物种 Species | 菌株 Strains | 分离地和基物 Isolation places and substrates | 产毒素 Toxin production | |
|---|---|---|---|---|---|
| AFB | CPA | ||||
| 36 | A. flavus | AB34 | 四川若尔盖; 土壤 Soil; Ruoergai Prairie, Sichuan, China | - | + |
| 37 | A. flavus | FJ17-2 | 福建宁德; 茶叶 Tea; Ningde, Fujian, China | + | + |
| 38 | A. flavus | HB4 | 湖北神农架; 土壤 Soil; Shennongjia, Hubei, China | + | + |
| 39 | A. flavus | HL53 | 黑龙江凉水; 土壤 Soil; Liangshui Nature Reserve, Heilongjiang, China | + | + |
| 40 | A. flavus | HL70 | 黑龙江乌伊岭; 土壤 Soil; Wuyiling, Heilongjiang, China | + | + |
| 41 | A. flavus | KK39 | 青海互助县; 土壤 Soil; Huzhu County, Qinghai, China | - | + |
| 42 | A. flavus | KK41 | 青海互助县; 土壤 Soil; Huzhu County, Qinghai, China | - | + |
| 43 | A. flavus | KK49 | 青海互助县; 土壤 Soil; Huzhu County, Qinghai, China | - | + |
| 44 | A. flavus | KK50 | 青海可可西里; 土壤 Soil; Hoh Xil, Qinghai, China | - | + |
| 45 | A. flavus | KK65 | 青海楚玛尔河; 土壤 Soil; Chumaer River, Qinghai, China | - | + |
| 46 | A. flavus | KK66 | 青海可可西里; 土壤 Soil; Hoh Xil, Qinghai, China | - | + |
| 47 | A. flavus | KK67 | 青海楚玛尔河; 土壤 Soil; Chumaer River, Qinghai, China | - | + |
| 48 | A. flavus | KK68 | 青海沱沱河; 土壤 Soil; Tuotuo River, Qinghai, China | - | + |
| 49 | A. flavus | KK69 | 青海沱沱河; 土壤 Soil; Tuotuo River, Qinghai, China | - | + |
| 50 | A. flavus | KK70 | 青海楚玛尔河; 土壤 Soil; Chumaer River, Qinghai, China | - | + |
| 51 | A. flavus | KK72 | 青海青海湖; 土壤 Soil; Qinghai Lake, Qinghai, China | + | + |
| 52 | A. flavus | KK73 | 青海青海湖; 土壤 Soil; Qinghai Lake, Qinghai, China | + | + |
| 53 | A. flavus | KK94 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | + | + |
| 54 | A. flavus | KK102 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 55 | A. flavus | KK103 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 56 | A. flavus | KK104 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 57 | A. flavus | KK114 | 青海坎布拉; 土壤 Soil; Kanbula, Qinghai, China | - | + |
| 58 | A. flavus | XZ107 | 陕西南宫山; 植物叶 Plant leaves; Mt. Nangongshan, Shaanxi, China | + | + |
| 59 | A. flavus | XZ108 | 陕西通天河; 植物叶 Plant leaves; Tongtian River, Shaanxi, China | + | + |
| 60 | A. flavus | XZ109 | 陕西南宫山; 植物叶 Plant leaves; Mt. Nangongshan, Shaanxi, China | + | + |
| 61 | A. flavus | XZ112 | 陕西通天河; 植物叶 Plant leaves; Tongtian River, Shaanxi, China | + | + |
| 62 | A. flavus | YN23 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | - | + |
| 63 | A. flavus | YN35 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 64 | A. flavus | YN48 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 65 | A. flavus | YN49 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 66 | A. flavus | YN51 | 云南玉溪; 烟叶 Tobacco leaves; Yuxi, Yunnan, China | + | + |
| 67 | A. flavus | NRRL 3357 | 美国; 霉花生 Moldy peanuts; USA | + | + |
| 68 | A. oryzae | RIB40 | 日本; 谷粒 Cereal grains; Japan | - | - |
| 69 | A. flavus | 3.262 | 辽宁大连; 空气 Air; Dalian, Liaoning, China | - | - |
| 70 | A. flavus | 3.267 | 辽宁大连; 土壤 Soil; Dalian, Liaoning, China | - | + |
| 71 | A. flavus | 3.337 | 天津; 蚊香 Mosquito-repellent incense; Tianjin, China | - | + |
| 72 | A. flavus | 3.417 | 天津; 酱曲 Soy sauce starter; Tianjin, China | - | + |
| 73 | A. flavus | 3.870 | 天津; 酱曲 Soy sauce starter; Tianjin, China | + | + |
| 74 | A. flavus | 3.881 | 上海; 小麦 Wheat; Shanghai, China | - | + |
| 75 | A. flavus | 3.2146 | 北京; 大米 Rice; Beijing, China | + | + |
| 76 | A. flavus | 3.2758 | 广东广州; 空气 Air; Guangzhou, Guangdong, China | - | + |
| 77 | A. flavus | 3.2789 | 越南河内; 土壤 Soil; Hanoi, Vietnam | - | + |
| 78 | A. flavus | 3.2823 | 安徽芜湖; 植物 Plants; Wuhu, Anhui, China | - | + |
| 菌株顺序号* Number | 物种 Species | 菌株 Strains | 分离地和基物 Isolation places and substrates | 产毒素 Toxin production | |
|---|---|---|---|---|---|
| AFB | CPA | ||||
| 79 | A. flavus | 3.3554 | 北京; 空气 Air; Beijing, China | - | + |
| 80 | A. flavus var. columnaris | CBS 485.65T | 日本; 黄油 Ex-type of A. flavus var. columnaris, butter; Japan | - | + |
| 81 | A. flavus | 3.4408-2 | 北京; 空气 Air; Beijing, China | + | + |
| 82 | A. flavus | 3.4410 | 美国 ATCC 28539; USA | + | + |
| 83 | A. flavus | 3.5211 | 北京 CICC 2348; Beijing, China | - | + |
| 84 | A. flavus | 3.5278 | 四川德阳; 烂水果 Rotten fruit; Deyang, Sichuan, China | + | + |
| 85 | A. flavus | 3.5283 | 四川成都; 土壤 Soil; Chengdu, Sichuan, China | + | + |
| 86 | A. flavus | 3.5309 | 四川都江堰; 土壤 Soil; Dujiangyan, Sichuan, China | + | + |
| 87 | A. flavus | 3.5329 | 贵州梵净山; 皮革 Leather; Mt. Fanjingshan, Guizhou, China | + | + |
| 88 | A. flavus | 3.6153 | 山东泰安; 小麦 Wheat; Tai’an, Shandong, China | - | + |
| 89 | A. flavus | 3.6304 | 广西宜山; 玉米 Corn; Yishan, Guangxi, China | - | + |
| 90 | A. flavus | 3.6307 | 吉林珲春; 亚麻 Linen; Hunchun, Jilin, China | + | + |
| 91 | A. flavus | 3.6311 | 广东广州; 空气 Air; Guangzhou, Guangdong, China | + | + |
| 92 | A. flavus | 3.6422 | 河北小五台山; 松果 Pinecore; Mt. Small Wutaishan, Hebei, China | + | + |
| 93 | A. flavus | 3.6428 | 云南大理; 霉纸 Mouldy paper; Dali, Yunnan, China | + | + |
| 94 | A. flavus | 3.6431 | 云南大理; 玉米叶 Corn leaves; Dali, Yunnan, China | + | + |
| 95 | A. flavus | 3.6434 | 云南思茅; 土壤 Soil; Simao, Yunnan, China | - | + |
| 96 | A. flavus | 14160 | 河南信阳; 土壤 Soil,; Xinyang, Henan, China | - | - |
| 97 | A. flavus | FJ17 | 福建宁德; 茶叶 Tea; Ningde, Fujian, China | + | + |
| A. arachidicola | CBS 117610T | 阿根廷; 花生叶 Arachis glabrata leaves; Argentina | |||
| A. minisclerotigenes | CBS 115635T | 阿根廷; 花生 Arachis hypogaea seeds; Argentina | |||
| A. parasiticus | CBS 100926T | 美国夏威夷; 嗜桔粉蚧 Pseudococcus calceolariae; Hawaii, USA | |||
| 3.306 | 天津; 酱曲 Soy sauce starter; Tianjin, China | ||||
| A. aflatoxiformans | CBS 143679T | 尼日利亚; 土壤 Soil; Nigeria | |||
| A. tamarii | CBS 104.13T | 分离地未知; 活性碳 Activated carbon; unknown country | |||
Table 1 (continued)
| 菌株顺序号* Number | 物种 Species | 菌株 Strains | 分离地和基物 Isolation places and substrates | 产毒素 Toxin production | |
|---|---|---|---|---|---|
| AFB | CPA | ||||
| 79 | A. flavus | 3.3554 | 北京; 空气 Air; Beijing, China | - | + |
| 80 | A. flavus var. columnaris | CBS 485.65T | 日本; 黄油 Ex-type of A. flavus var. columnaris, butter; Japan | - | + |
| 81 | A. flavus | 3.4408-2 | 北京; 空气 Air; Beijing, China | + | + |
| 82 | A. flavus | 3.4410 | 美国 ATCC 28539; USA | + | + |
| 83 | A. flavus | 3.5211 | 北京 CICC 2348; Beijing, China | - | + |
| 84 | A. flavus | 3.5278 | 四川德阳; 烂水果 Rotten fruit; Deyang, Sichuan, China | + | + |
| 85 | A. flavus | 3.5283 | 四川成都; 土壤 Soil; Chengdu, Sichuan, China | + | + |
| 86 | A. flavus | 3.5309 | 四川都江堰; 土壤 Soil; Dujiangyan, Sichuan, China | + | + |
| 87 | A. flavus | 3.5329 | 贵州梵净山; 皮革 Leather; Mt. Fanjingshan, Guizhou, China | + | + |
| 88 | A. flavus | 3.6153 | 山东泰安; 小麦 Wheat; Tai’an, Shandong, China | - | + |
| 89 | A. flavus | 3.6304 | 广西宜山; 玉米 Corn; Yishan, Guangxi, China | - | + |
| 90 | A. flavus | 3.6307 | 吉林珲春; 亚麻 Linen; Hunchun, Jilin, China | + | + |
| 91 | A. flavus | 3.6311 | 广东广州; 空气 Air; Guangzhou, Guangdong, China | + | + |
| 92 | A. flavus | 3.6422 | 河北小五台山; 松果 Pinecore; Mt. Small Wutaishan, Hebei, China | + | + |
| 93 | A. flavus | 3.6428 | 云南大理; 霉纸 Mouldy paper; Dali, Yunnan, China | + | + |
| 94 | A. flavus | 3.6431 | 云南大理; 玉米叶 Corn leaves; Dali, Yunnan, China | + | + |
| 95 | A. flavus | 3.6434 | 云南思茅; 土壤 Soil; Simao, Yunnan, China | - | + |
| 96 | A. flavus | 14160 | 河南信阳; 土壤 Soil,; Xinyang, Henan, China | - | - |
| 97 | A. flavus | FJ17 | 福建宁德; 茶叶 Tea; Ningde, Fujian, China | + | + |
| A. arachidicola | CBS 117610T | 阿根廷; 花生叶 Arachis glabrata leaves; Argentina | |||
| A. minisclerotigenes | CBS 115635T | 阿根廷; 花生 Arachis hypogaea seeds; Argentina | |||
| A. parasiticus | CBS 100926T | 美国夏威夷; 嗜桔粉蚧 Pseudococcus calceolariae; Hawaii, USA | |||
| 3.306 | 天津; 酱曲 Soy sauce starter; Tianjin, China | ||||
| A. aflatoxiformans | CBS 143679T | 尼日利亚; 土壤 Soil; Nigeria | |||
| A. tamarii | CBS 104.13T | 分离地未知; 活性碳 Activated carbon; unknown country | |||
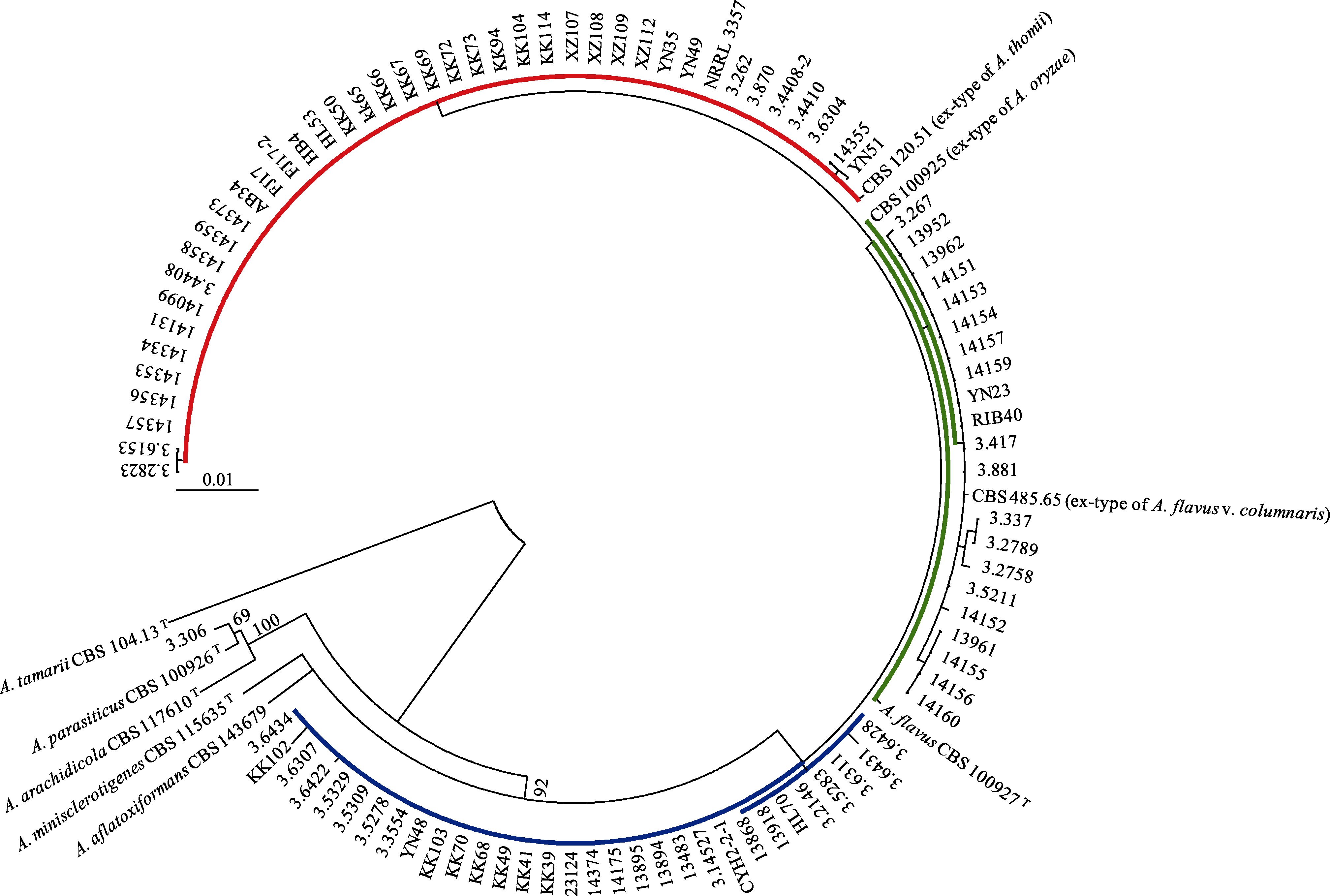
Fig. 1 The maximum likelihood phylogram of 97 Aspergillus flavus strains and its four close-related species. The ex-type of A. flavus CBS 100927T, ex-type of A. flavus var. columnaris CBS485.65T, ex-type of A. oryzae CBS 100925T, and ex-type of A. thomii CBS 120.51T are in the same clade with a 92% support, with the ex-type of A. tamari CBS 104.13T as the outgroup. The red, green and blue colours are in accordance with Fig. 3.
| K | Replicates | Mean LnP(K) | Stdev LnP(K) | Ln°(K) | |Ln?(K)| | Delta K |
|---|---|---|---|---|---|---|
| 2 | 20 | -252.900000 | 11.116323 | - | - | - |
| 3 | 20 | -153.585000 | 0.665918 | 99.315000 | 115.770000 | 173.850119 |
| 4 | 20 | -170.040000 | 4.371607 | -16.455000 | 14.975000 | 3.425514 |
| 5 | 20 | -171.520000 | 3.293790 | -1.480000 | 16.860000 | 5.118723 |
| 6 | 20 | -189.860000 | 14.895474 | -18.340000 | 402.695000 | 27.034721 |
| 7 | 20 | -610.895000 | 1,768.728466 | -421.035000 | 779.785000 | 0.440873 |
| 8 | 20 | -252.145000 | 53.166892 | 358.750000 | — | — |
Table 2 The best population number K inferred by Structure 2.3.4
| K | Replicates | Mean LnP(K) | Stdev LnP(K) | Ln°(K) | |Ln?(K)| | Delta K |
|---|---|---|---|---|---|---|
| 2 | 20 | -252.900000 | 11.116323 | - | - | - |
| 3 | 20 | -153.585000 | 0.665918 | 99.315000 | 115.770000 | 173.850119 |
| 4 | 20 | -170.040000 | 4.371607 | -16.455000 | 14.975000 | 3.425514 |
| 5 | 20 | -171.520000 | 3.293790 | -1.480000 | 16.860000 | 5.118723 |
| 6 | 20 | -189.860000 | 14.895474 | -18.340000 | 402.695000 | 27.034721 |
| 7 | 20 | -610.895000 | 1,768.728466 | -421.035000 | 779.785000 | 0.440873 |
| 8 | 20 | -252.145000 | 53.166892 | 358.750000 | — | — |
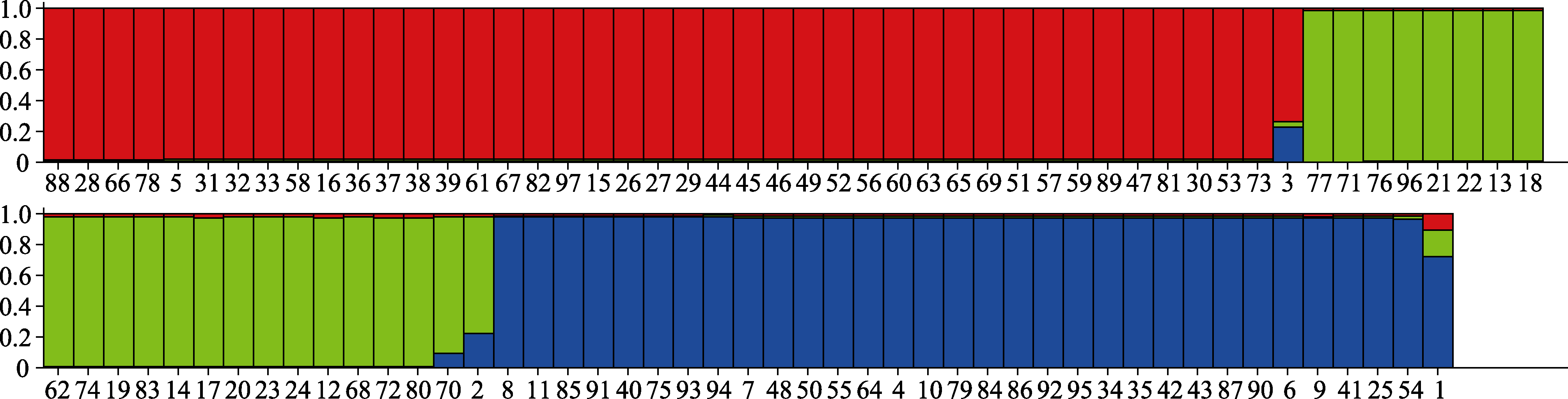
Fig. 3 The three populations of the 97 Aspergillus flavus isolates calculated by Structure Harvester. Red, green and blue colours stand for the three different populations, each column stands for each strain, and the number under each column is in accordance with those in Table 1.
| 1 | Anderson B, Thrane U ( 2006) Food-borne fungi in fruit and cereals and their production of mycotoxins. In: Advances in Food Mycology (eds Hocking AD, Pitt JI, Samson RA, Thrane U), pp. 137-152. Springer, Boston. |
| 2 | Barros G, Torres A, Chulze S ( 2005) Aspergillus flavus population isolated from soil of Argentina’s peanut-growing region. Sclerotia production and toxigenic profile. Journal of the Science of Food and Agriculture, 85, 2349-2353. |
| 3 | Batista PP, Santos JF, Oliveira NT, Pires APD, Motta CMS, Luna-Alves Lima EA ( 2008) Genetic characterization of Brazilian strains of Aspergillus flavus using DNA markers. Genetics and Molecular Research, 7, 706-717. |
| 4 | CAST ( Council for Agricultural Science and Technology) ( 2003) Mycotoxins: Risks in plant, animal and human systems. Task Force Report No. 139, 13-85. CAST, Ames, IA. |
| 5 | Chang PK, Ehrlich KC ( 2010) What does genetic diversity of Aspergillus flavus tell us about Aspergillus flavus? International Journal of Food Microbiology, 138, 189-199. |
| 6 | Chang PK, Ehrlich KC, Hua SS ( 2006) Cladal relatedness among Aspergillus flavus isolates and Aspergillus flavus S and L morphotype isolates. International Journal of Food Microbiology, 108, 172-177. |
| 7 | Cotty PJ ( 1997) Aflatoxin-producing potential of communities of Aspergillus section Flavi from cotton producing areas in the United States. Mycological Research, 101, 698-704. |
| 8 | Cotty PJ ( 1989) Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology, 79, 808-814. |
| 9 | Cotty PJ, Jaime-Garcia R ( 2007) Influence of climate on aflatoxin producing fungi and aflatoxin contamination. International Journal of Food Microbiology, 119, 109-115. |
| 10 | Ehrlich KC, Chang P-K, Yu J, Cotty PJ ( 2004) Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Applied and Environmental Microbiology, 70, 6518-6524. |
| 11 | Frisvad JC, Skouboe P, Samson RA ( 2005) Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylstergmatocystin, Aspergillus rambellii sp. nov. Systematic and Applied Microbiology, 28, 442-453. |
| 12 | Frisvad JC, Hubka V, Ezekiel CN, Hong SB, Novakova A, Chen AJ, Arzanlou M, Larsen TO, Sklenar F, Mahakarnchanakul W, Samson RA, Houbraken J ( 2019) Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology, 93, 1-63. |
| 13 | Gao XF, Yin SA, Ji R ( 2011) Contamination of aflatoxins in peanuts from some regions in China. Chinese Journal of Public Health, 27, 541-542. |
| (in Chinese with English abstract) [ 高秀芬, 荫士安, 计融 ( 2011) 中国部分地区花生中4种黄曲霉毒素污染调查. 中国公共卫生, 27, 541-542.] | |
| 14 | Geiser DM, Dorner JW, Horn BW, Taylor JW ( 2000) The phylogenetics of mycotoxin and sclertium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genetics and Biology, 31, 169-179. |
| 15 | Glass NL, Donaldson GC ( 1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology, 61, 1323-1330. |
| 16 | Hall TA ( 1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| 17 | Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW ( 2007) Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology, 153, 1677-1692. |
| 18 | Horn BW, Greene RL, Dorner JW ( 1995) Effect of corn and peanut cultivation on soil populations of Aspergillus flavus and A. parasiticus in southwestern Georgia. Applied and Environmental Microbiology, 61, 2472-2475. |
| 19 | Horn BW, Dorner JW ( 1999) Regional differences in production of aflatoxin B1 and cyclopiazonic acid by soil isolates of Aspergillus flavus along a transect within the United States. Applied and Environmental Microbiology, 65, 1444-1449. |
| 20 | Horn BW ( 2003) Ecology and population biology of aflatoxingenic fungi in soil. Journal of Toxicology-Toxin Reviews, 22, 351-379. |
| 21 | Krishnan S, Manavathu EK, Chandrasekar PH , ( 2009) Aspergillus flavus: An emerging non-fumigatus Aspergillus species of significance. Mycoses, 52, 206-222. |
| 22 | Kurtzman CP, Smiley MJ, Robnett CJ, Wicklow DT ( 1986) DNA relatedness among wild and domesticated species in the Aspergillus flavus Group. Mycologi, 78, 955-959. |
| 23 | Malloch D ( 1981) Moulds Their Isolation, Cultivation and Identification. University of Toronto Press, Toronto. |
| 24 | Medina A, Gilbert MK, Mack BM, Brian GR, Rodríguez A, Bhatnagar D, Payne G, Magan N ( 2017) Interactions between water activity and temperature on the Aspergillus flavus transcriptome and aflatoxin B1 production. International Journal of Food Microbiology, 256, 36-44. |
| 25 | Orum TV, Bigelow DM, Nelson MR, Howell DR, Cotty PJ ( 1997) Spatial and temporal patterns of Aspergillus flavus strain composition and propagule density in Yuma County, Arizona, soils. Plant Diseases, 81, 911-916. |
| 26 | Paterson RRM, Lima N ( 2010) How will climate change affect mycotoxins in food? Food Research International, 43, 1902-1914. |
| 27 | Pildain MB, Frisvad JC, Vaamonde G, Cabral D, Varga J, Samson RA ( 2008) Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. International Journal of Systematic and Evolutionary Microbiology, 58, 725-735. |
| 28 | Pitt JI, Hocking AD ( 2009) Fungi and Food spoilage, 3rd edn. Springer-Science Media, London. |
| 29 | Qi ZT, Kong HZ, Sun ZM ( 1997) Flora Fungorum Sinicorum Vol. 5. Aspergillus et teleomorphi cognate. Science Press, Beijing. |
| ( In Chinese) [ 齐祖同, 孔华忠, 孙曾美( 1997) 中国真菌志第五卷: 曲霉属及其相关有性型. 科学出版社, 北京.] | |
| 30 | Raper KB, Fennell DI ( 1965) The Genus Aspergillus. Williams & Wilkins, Baltimore. |
| 31 | Saito M, Tsuruta O ( 1993) A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. Proceedings of the Japanese Association of Mycotoxicology, 37, 31-36. |
| 32 | Sepahvand A, Shams-Ghahfarokhi M, Allameh A, Jahanshiri Z, Jamali M, Razzaghi-Abyaneh M ( 2011) A survey on distribution and toxigenicity of Aspergillus flavus from indoor and outdoor hospital environments. Folia Microbiologica, 56, 527-534. |
| 33 | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S ( 2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 8, 2731-2739. |
| 34 | Varga J, Frisvad JC, Samson RA ( 2011) Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Studies in Mycology, 69, 57-80. |
| 35 | Wang L, Zhuang WY ( 2004) Designing primer sets for amplification of partial calmodulin genes from penicillia. Mycosystema, 23, 466-473. |
| 36 | Wang L ( 2012) Four new records of Aspergillus section Usti from Shandong Province, China. Mycotaxon, 120, 373-384. |
| 37 | Wang ZG, Zhe T, Cheng SY, Cong LM ( 1993) Study of pectinase and sclerotium producing abilities of two kinds of Aspergillus flavus isolated from Zhejiang. Mycopathologia, 121, 163-168. |
| 38 | Wang HC, Huang YC, Wang J, Wang MS, Shang SH, Ye DY, Long MJ ( 2014) Fungi isolation and identification of tobacco seeds. Chinese Tobacco Science, 35(5), 84-88. |
| (in Chinese with English abstract) [ 汪汉成, 黄艳飞, 王进, 王茂胜, 商胜华, 叶定勇, 龙明锦 ( 2014) 烟草种子携带病原真菌的分离与鉴定. 中国烟草科学, 35(5), 84-88.] |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()