



Biodiv Sci ›› 2013, Vol. 21 ›› Issue (3): 315-325. DOI: 10.3724/SP.J.1003.2013.11035 cstr: 32101.14.SP.J.1003.2013.11035
• Orginal Article • Previous Articles Next Articles
Zhouhe Du1,2, Junfeng Liu1,*, Binbin Liu1, Yanchun Zuo1, Jianmei Wu1, Yi-an Chen1, Jianfei Zhang1, Cheng Lu2,*( )
)
Received:2013-02-04
Accepted:2013-04-26
Online:2013-05-20
Published:2013-06-05
Contact:
Liu Junfeng,Lu Cheng
Zhouhe Du,Junfeng Liu,Binbin Liu,Yanchun Zuo,Jianmei Wu,Yi-an Chen,Jianfei Zhang,Cheng Lu. Genetic diversity and molecular genealogy of local silkworm varieties[J]. Biodiv Sci, 2013, 21(3): 315-325.
| 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype | 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype |
|---|---|---|---|---|---|---|---|
| 1 | 小石丸 Xiaoshiwan | XSW | CV1 | 37 | 萨尼斯 Sanisi | SNS | EV1 |
| 2 | 二毛 Ermao | EM | CV1 | 38 | 罗尼7 Luoni7 | LN7 | EV1 |
| 3 | 土三虎斑 Tusanhuban | TSHB | CV1 | 39 | 法408 Fa408 | F408 | EV1 |
| 4 | 中土 Zhongtu | ZT | CV1 | 40 | 欧18 Ou18 | O18 | EV1 |
| 5 | 达县土种 Daxiantuzhong | DXTZ | CV1 | 41 | 意16 Yi16 | Y16 | EV1 |
| 6 | 土一肉黄圆 Tuyirouhuangyuan | TYRHY | CV1 | 42 | 乌D WuD | WD | EV2 |
| 7 | 土八 Tuba | Tba | CV1 | 43 | 乌G WuG | WG | EV2 |
| 8 | 鲁1 Lu1 | L1 | CV1 | 44 | 乌B WuB | WB | EV2 |
| 9 | Re黑 Rehei | RH | CV1 | 45 | 乌2 Wu2 | W2 | EV2 |
| 10 | 鲁10 Lu10 | L10 | CV1 | 46 | 日110 Ri110 | R110 | T |
| 11 | 土种01 Tuzhong01 | TZ01 | CV1 | 47 | 大造 Dazao | DZ | T |
| 12 | 诸桂 Zhugui | ZG | CV1 | 48 | 高花 Gaohua | GH | T |
| 13 | 温州种 Wenzhouzhong | WZZ | CV1 | 49 | 第五白卵 Diwubailuan | DWBL | T |
| 14 | 芙蓉灰卵 Furonghuiluan | FRHL | CV1 | 50 | 演金黄白 Yanjinhuangbai | YJHB | T |
| 15 | 灰色卵 Huiseluan | HSL | CV1 | 51 | 15-100 | 15-100 | T |
| 16 | 盱眙种 Xuyizhong | XYZ | CV1 | 52 | 07-310 | 07-310 | T |
| 17 | 余杭24 Yuhang24 | YH24 | CV1 | 53 | 高白 Gaobai | GB | T |
| 18 | 沛县种 Peixianzhong | PXZ | CV1 | 54 | 琼山海南 Qiongshanhainan | QSHN | T |
| 19 | 邯郸种 Handanzhong | HDZ | CV1 | 55 | 655 | 655 | T |
| 20 | 中4010 Zhong4010 | Z4010 | CV2 | 56 | 防四 Fangsi | FS | T |
| 21 | 泰 Tai | T | CV2 | 57 | 印度种 Yinduzhong | YDZ | T |
| 22 | 玉溪2号 Yuxi2hao | YX | CV2 | 58 | 加秋 Jiaqiu | JQ | T |
| 23 | 平 Ping | P | CV2 | 59 | 白夏B BaixiaB | BXB | T |
| 24 | C108 | C108 | CV2 | 60 | 天龙青白 Tianlongqingbai | TLQB | JV1 |
| 25 | 余杭2 Yuhang2 | YH2 | CV2 | 61 | 赤熟 Chishu | Cshu | JV1 |
| 26 | 土白 Tubai | Tbai | CV2 | 62 | 日9 Ri9 | R9 | JV1 |
| 27 | 松花形吴 Songhuaxingwu | SHXW | CV2 | 63 | 姬蚕 Jican | JC | JV1 |
| 28 | 保黄 Baohuang | BH | CVd | 64 | 黑子 Heizi | HZ | JV1 |
| 29 | MCH101 | MCH101 | CVd | 65 | 日10 Ri10 | R10 | JV1 |
| 30 | BT924 | BT924 | CVd | 66 | 绵蚕 Miancan | MC | JV1 |
| 31 | BH863 | BH863 | CVd | 67 | 日限2 Rixian2 | RX2 | JV2 |
| 表1 (续) Table 1 (continued) | |||||||
| 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype | 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype |
| 32 | 琼十 Qiongshi | QS | CVd | 68 | 赢纹形 Yingwenxing | YWX | JV2 |
| 33 | 四海 Sihai | SH | CVd | 69 | J115 | J115 | JV2 |
| 34 | 兰十 Lanshi | LanS | CVd | 70 | 830 | 830 | JV2 |
| 35 | 罗萨 Luosa | LuoS | EV1 | 71 | 春四 Chunsi | CS | JV2 |
| 36 | 阿利可斯 Alikesi | ALKS | EV1 | 72 | T8 | T8 | JV2 |
Table 1 The origin and characters of 72 silkworm varieties used in this study
| 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype | 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype |
|---|---|---|---|---|---|---|---|
| 1 | 小石丸 Xiaoshiwan | XSW | CV1 | 37 | 萨尼斯 Sanisi | SNS | EV1 |
| 2 | 二毛 Ermao | EM | CV1 | 38 | 罗尼7 Luoni7 | LN7 | EV1 |
| 3 | 土三虎斑 Tusanhuban | TSHB | CV1 | 39 | 法408 Fa408 | F408 | EV1 |
| 4 | 中土 Zhongtu | ZT | CV1 | 40 | 欧18 Ou18 | O18 | EV1 |
| 5 | 达县土种 Daxiantuzhong | DXTZ | CV1 | 41 | 意16 Yi16 | Y16 | EV1 |
| 6 | 土一肉黄圆 Tuyirouhuangyuan | TYRHY | CV1 | 42 | 乌D WuD | WD | EV2 |
| 7 | 土八 Tuba | Tba | CV1 | 43 | 乌G WuG | WG | EV2 |
| 8 | 鲁1 Lu1 | L1 | CV1 | 44 | 乌B WuB | WB | EV2 |
| 9 | Re黑 Rehei | RH | CV1 | 45 | 乌2 Wu2 | W2 | EV2 |
| 10 | 鲁10 Lu10 | L10 | CV1 | 46 | 日110 Ri110 | R110 | T |
| 11 | 土种01 Tuzhong01 | TZ01 | CV1 | 47 | 大造 Dazao | DZ | T |
| 12 | 诸桂 Zhugui | ZG | CV1 | 48 | 高花 Gaohua | GH | T |
| 13 | 温州种 Wenzhouzhong | WZZ | CV1 | 49 | 第五白卵 Diwubailuan | DWBL | T |
| 14 | 芙蓉灰卵 Furonghuiluan | FRHL | CV1 | 50 | 演金黄白 Yanjinhuangbai | YJHB | T |
| 15 | 灰色卵 Huiseluan | HSL | CV1 | 51 | 15-100 | 15-100 | T |
| 16 | 盱眙种 Xuyizhong | XYZ | CV1 | 52 | 07-310 | 07-310 | T |
| 17 | 余杭24 Yuhang24 | YH24 | CV1 | 53 | 高白 Gaobai | GB | T |
| 18 | 沛县种 Peixianzhong | PXZ | CV1 | 54 | 琼山海南 Qiongshanhainan | QSHN | T |
| 19 | 邯郸种 Handanzhong | HDZ | CV1 | 55 | 655 | 655 | T |
| 20 | 中4010 Zhong4010 | Z4010 | CV2 | 56 | 防四 Fangsi | FS | T |
| 21 | 泰 Tai | T | CV2 | 57 | 印度种 Yinduzhong | YDZ | T |
| 22 | 玉溪2号 Yuxi2hao | YX | CV2 | 58 | 加秋 Jiaqiu | JQ | T |
| 23 | 平 Ping | P | CV2 | 59 | 白夏B BaixiaB | BXB | T |
| 24 | C108 | C108 | CV2 | 60 | 天龙青白 Tianlongqingbai | TLQB | JV1 |
| 25 | 余杭2 Yuhang2 | YH2 | CV2 | 61 | 赤熟 Chishu | Cshu | JV1 |
| 26 | 土白 Tubai | Tbai | CV2 | 62 | 日9 Ri9 | R9 | JV1 |
| 27 | 松花形吴 Songhuaxingwu | SHXW | CV2 | 63 | 姬蚕 Jican | JC | JV1 |
| 28 | 保黄 Baohuang | BH | CVd | 64 | 黑子 Heizi | HZ | JV1 |
| 29 | MCH101 | MCH101 | CVd | 65 | 日10 Ri10 | R10 | JV1 |
| 30 | BT924 | BT924 | CVd | 66 | 绵蚕 Miancan | MC | JV1 |
| 31 | BH863 | BH863 | CVd | 67 | 日限2 Rixian2 | RX2 | JV2 |
| 表1 (续) Table 1 (continued) | |||||||
| 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype | 序号 No. | 品种 Variety | 代号 Code | 生态类型 Ecotype |
| 32 | 琼十 Qiongshi | QS | CVd | 68 | 赢纹形 Yingwenxing | YWX | JV2 |
| 33 | 四海 Sihai | SH | CVd | 69 | J115 | J115 | JV2 |
| 34 | 兰十 Lanshi | LanS | CVd | 70 | 830 | 830 | JV2 |
| 35 | 罗萨 Luosa | LuoS | EV1 | 71 | 春四 Chunsi | CS | JV2 |
| 36 | 阿利可斯 Alikesi | ALKS | EV1 | 72 | T8 | T8 | JV2 |
| 序列长度 Sequence length (bp) | 碱基插入/缺失位点数 Insert/delete (%) | 变异位点数 Variable site (%) | 单个突变位点 Singleton variable site (%) | 简约信息位点 Parsimony variable site (%) | |
|---|---|---|---|---|---|
| 外显子 Exon | 493 | 2(0.4)* | 10(2.0) | 2(0.4) | 8(1.6) |
| 内含子 Intron | 670 | 139(20.7) | 55(8.2) | 7(1.0) | 48(7.2) |
| 全序列 Entire | 1,163 | 141(12.1) | 65(5.6) | 9(0.8) | 56(4.8) |
Table 2 Statistics of the varieties bases of 72 local silkworm varieties in the Bmamy2 gene amplified region
| 序列长度 Sequence length (bp) | 碱基插入/缺失位点数 Insert/delete (%) | 变异位点数 Variable site (%) | 单个突变位点 Singleton variable site (%) | 简约信息位点 Parsimony variable site (%) | |
|---|---|---|---|---|---|
| 外显子 Exon | 493 | 2(0.4)* | 10(2.0) | 2(0.4) | 8(1.6) |
| 内含子 Intron | 670 | 139(20.7) | 55(8.2) | 7(1.0) | 48(7.2) |
| 全序列 Entire | 1,163 | 141(12.1) | 65(5.6) | 9(0.8) | 56(4.8) |
| CV2 | EV1 | T | CV1M3 | JV2 | CV1M4 | JV1 | CVd | |
|---|---|---|---|---|---|---|---|---|
| CV2 | ||||||||
| EV1 | 0.025 | |||||||
| T | 0.027 | 0.024 | ||||||
| CV1M3 | 0.026 | 0.015 | 0.023 | |||||
| JV2 | 0.026 | 0.012 | 0.024 | 0.016 | ||||
| CV1M4 | 0.026 | 0.013 | 0.024 | 0.017 | 0.014 | |||
| JV1 | 0.027 | 0.025 | 0.024 | 0.025 | 0.026 | 0.025 | ||
| CVd | 0.025 | 0.025 | 0.024 | 0.025 | 0.026 | 0.025 | 0.024 | |
| EV2 | 0.025 | 0.044 | 0.029 | 0.039 | 0.044 | 0.042 | 0.026 | 0.026 |
Table 3 Genetic distance among different ecotypic varieties of local silkworm
| CV2 | EV1 | T | CV1M3 | JV2 | CV1M4 | JV1 | CVd | |
|---|---|---|---|---|---|---|---|---|
| CV2 | ||||||||
| EV1 | 0.025 | |||||||
| T | 0.027 | 0.024 | ||||||
| CV1M3 | 0.026 | 0.015 | 0.023 | |||||
| JV2 | 0.026 | 0.012 | 0.024 | 0.016 | ||||
| CV1M4 | 0.026 | 0.013 | 0.024 | 0.017 | 0.014 | |||
| JV1 | 0.027 | 0.025 | 0.024 | 0.025 | 0.026 | 0.025 | ||
| CVd | 0.025 | 0.025 | 0.024 | 0.025 | 0.026 | 0.025 | 0.024 | |
| EV2 | 0.025 | 0.044 | 0.029 | 0.039 | 0.044 | 0.042 | 0.026 | 0.026 |
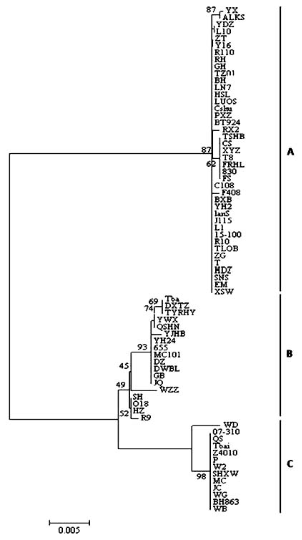
Fig. 4 Phylogenetic (NJ) tree based on nucleotide sequence of Bmamy2 gene of Bombyx mori. Nucleotide P-distance model and pairwise deletion of gaps were selected for the tree reconstruction in the program MEGA 4.0, and bootstrap values (1,000 replicate) of >40% are shown. Variety codes see Table 1.
| 聚类支 Clade | 地理生态类型 Ecotype | 品种数(个) No. of varieties | 占各支百分比 % |
|---|---|---|---|
| A | 中系一化 CV1 | 15 | 36.59 |
| 中系二化 CV2 | 4 | 9.76 | |
| 中系多化 CVd | 3 | 7.32 | |
| 日系一化 JV1 | 3 | 7.32 | |
| 日系二化 JV2 | 5 | 12.20 | |
| 欧系一化 EV1 | 6 | 14.63 | |
| 热带系统 T | 5 | 12.20 | |
| 小计 Sum | 41 | ||
| B | 中系一化 CV1 | 6 | 33.33 |
| 中系多化 CVd | 2 | 11.11 | |
| 日系一化 JV1 | 2 | 11.11 | |
| 日系二化 JV2 | 1 | 5.56 | |
| 欧系一化 EV1 | 1 | 5.46 | |
| 热带系统 T | 6 | 33.33 | |
| 小计 Sum | 18 | ||
| C | 中系二化 CV2 | 4 | 30.77 |
| 中系多化 CVd | 2 | 15.38 | |
| 日系一化 JV1 | 2 | 14.38 | |
| 欧系二化 EV2 | 4 | 30.77 | |
| 热带系统 T | 1 | 7.69 | |
| 小计 Sum | 13 | ||
| 合计 | Total | 72 |
Table 4 Silkworm varieties belonging to different clades
| 聚类支 Clade | 地理生态类型 Ecotype | 品种数(个) No. of varieties | 占各支百分比 % |
|---|---|---|---|
| A | 中系一化 CV1 | 15 | 36.59 |
| 中系二化 CV2 | 4 | 9.76 | |
| 中系多化 CVd | 3 | 7.32 | |
| 日系一化 JV1 | 3 | 7.32 | |
| 日系二化 JV2 | 5 | 12.20 | |
| 欧系一化 EV1 | 6 | 14.63 | |
| 热带系统 T | 5 | 12.20 | |
| 小计 Sum | 41 | ||
| B | 中系一化 CV1 | 6 | 33.33 |
| 中系多化 CVd | 2 | 11.11 | |
| 日系一化 JV1 | 2 | 11.11 | |
| 日系二化 JV2 | 1 | 5.56 | |
| 欧系一化 EV1 | 1 | 5.46 | |
| 热带系统 T | 6 | 33.33 | |
| 小计 Sum | 18 | ||
| C | 中系二化 CV2 | 4 | 30.77 |
| 中系多化 CVd | 2 | 15.38 | |
| 日系一化 JV1 | 2 | 14.38 | |
| 欧系二化 EV2 | 4 | 30.77 | |
| 热带系统 T | 1 | 7.69 | |
| 小计 Sum | 13 | ||
| 合计 | Total | 72 |
| 变异来源 Source of variance | 自由度 df | 方差分量 Variance component | 变异百分比 (%) | P-value |
|---|---|---|---|---|
| 地理生态类型组群间 Among geo-voltings | 7 | 6.93028 | 19.86 | > 0.05 |
| 地理生态类型组群内 Among population within geo-voltings | 1 | 1.23267 | 3.53 | > 0.05 |
| 种群内 Within populations | 63 | 26.73978 | 76.61 | < 0.01 |
Table 5 AMOVA of Bmamy2 gene for 72 silkworm varieties of Bombyx mori based on geo-ecotype
| 变异来源 Source of variance | 自由度 df | 方差分量 Variance component | 变异百分比 (%) | P-value |
|---|---|---|---|---|
| 地理生态类型组群间 Among geo-voltings | 7 | 6.93028 | 19.86 | > 0.05 |
| 地理生态类型组群内 Among population within geo-voltings | 1 | 1.23267 | 3.53 | > 0.05 |
| 种群内 Within populations | 63 | 26.73978 | 76.61 | < 0.01 |
| 1 | Arunkumar KP, Muralidhar M, Nagaraju J (2006) Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mori from Chinese Bombyx mandarina and paternal inheritance of Antheraea proylei mitochondrial DNA. Molecular Phylogenetics and Evolution, 40, 419-427. |
| 2 | Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| 3 | Brown WM (1983) Evolution of animal mitochondrial DNA. In: Evolution of Genes and Proteins (eds Nei M, Koehned RK), pp. 62-88. Sinauer Associates, Sunderland MA. |
| 4 | Chen LY (陈丽媛), Zhao QL (赵巧玲), Shen XJ (沈兴家), Zhang ZF (张志芳), Tang SM (唐顺明), Xu AY (徐安英), Zhang GZ (张国政), Guo XJ (郭锡杰) (2007) Nucleotide sequences in the A+T rich regions of mitochondrial DNA from local races of Bombyx mori and their molecular evolution. Science of Sericulture(蚕业科学), 33, 5-13. (in Chinese with English abstract) |
| 5 | Cheng XY (成新跃), Zhou HZ (周红章), Zhang GX (张广学) (2000) Perspective of molecular biological techniques applied in insect systematics. Acta Zootaxonomica Sinica(动物分类学报), 25, 121-133. (in Chinese with English abstract) |
| 6 | Du ZH (杜周和) (2009) The Origin and Differentiation of Silkworm, Bombyx mori, Based on the Bmamy2 Gene (基于Bmamy2基因的家蚕起源与分化研究), PhD dissertation, Southwest University, Chongqing. (in Chinese) |
| 7 | Du ZH (杜周和), Dong ZP (董占鹏), Liu JF (刘俊凤), Liu BB (刘彬斌), Lu C (鲁成) (2008) Cloning, expression pattern and sequence analysis of Bmamy2 gene in the silkwom, Bombyx mori. Science of Sericulture(蚕业科学), 34, 642-649. (in Chinese with English abstract) |
| 8 | Du ZH (杜周和), Liu JF (刘俊凤), Liu BB (刘彬斌), Dong ZP (董占鹏), Yu QY (余泉友), Lu C (鲁成), Chen YA (陈义安) (2009) Genetic diversity of the wild mulberry silkworm, Bombyx mandarina, in China and its phylogenetic relationship with the domesticated silkworm, Bombyx mori, based on amy gene. Acta Entomologica Sinica(昆虫学报), 52, 1338-1348. (in Chinese with English abstract) |
| 9 | Excoffier L, Smouse PE, Quattor JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491. |
| 10 | Fang SM (房守敏), Yu QY (余泉友), Li B (李斌), Dai FY (代方银), Miao XX (苗雪霞), Huang YP (黄勇平), Lu C (鲁成) (2007) Comparison of SSR, AFLP and RAPD markers for genetic analysis of domestic silkworm. Scientia Agricultura Sinica(中国农业科学), 40, 2926-2930. (in Chinese with English abstract) |
| 11 | Fang SM (房守敏), Zhang L (张烈), Lu C (鲁成) (2010) Study and application in origin and evolution of mitochondrial DNA in the silkmoths. Chinese Bulletin of Entomology(昆虫知识), 47, 439-445. (in Chinese with English abstract) |
| 12 | Gyllensten U, Wharton D, Josefsson A, Wilson AC (1991) Paternal inheritance of mitochondrial DNA in mice. Nature, 352, 255-257. |
| 13 | Hu XL, Cao GL, Xue RY, Zheng XJ, Zhang X, Duan HR, Gong CL (2010) The complete mitogenome and phylogenetic analysis of Bombyx mandarina variety Qingzhou. Molecular Biology Report, 37, 2599-2608. |
| 14 | Jiang YL(蒋猷龙) (1982) The Origin of Silkworm, Bombyx mori, and Its Differentiation(家蚕的起源与分化). Jiangsu Science and Technology Press, Nanjing(in Chinese) |
| 15 | Jiang YL (蒋猷龙) (1989) Advance of the study on the origin of silkworm and its differentiation. Newsletter of Agriculture and Animal Husbandry(农牧情报研究), (7), 43-47. (in Chinese) |
| 16 | Kenji Y, Hideki S, Toshiki T, Eiichi K, Makoto K (2011) Nucleotide sequence variation in mitochondrial COI gene among 147 silkworm (Bombyx mori) varieties from Japanese, Chinese, European and moltinism classes. Genes & Genetic Systems, 86, 315-323. |
| 17 | Li AL, Zhao QL, Tang SM, Zhang ZF, Pan SY, Shen GF (2005) Molecular phylogeny of the domesticated silkworm, Bombyx mori, based on the sequences of mitochondrial cytochrome b genes. Journal of Genetics, 84, 137-142. |
| 18 | Li D, Guo YR, Shao HJ, Tellier LC, Wang J, Xiang ZH, Xia QY (2010) Genetic diversity, molecular phylogeny and selection evidence of the silkworm mitochondria implicated by complete resequencing of 41 genomes. BMC Evolutionary Biology, 10, 81. |
| 19 | Li HW (李焕文) (1985) Exploration of the origin and voltinism change of silkworm, Bombyx mori, and mulberry silkworm, Bombyx mandarina. Newsletter of Sericultural Science(蚕学通讯), (1), 41-45. (in Chinese) |
| 20 | Li MW (李木旺) (2005) The Application of Microsatellite Markers in Silkworm (Bombyx mori): Genetic Diversity, QTL Analysis, Gene Mapping and Marker Assisted Selection (微卫星遗传标记在家蚕中的应用研究——遗传多样性分析、QTL分析、基因定位与分子标记辅助育种). PhD dissertation, Institute of Plant Physiology & Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai. (in Chinese) |
| 21 | Lu C (鲁成), Yu HS (余红仕), Xiang ZH (向仲怀) (2002a) Molecular systematic studies on Chinese mandarina silkworm (Bombyx mandarina M.) and domestic silkworm (Bombyx mori L.). Scientia Agricultura Sinica(中国农业科学), 35, 94-101. (in Chinese with English abstract) |
| 22 | Lu C (鲁成), Zhao AC (赵爱春), Zhou ZY (周泽扬), Xiang ZH (向仲怀), Wan CL (万春玲) (2001) AFLP analysis of Chinese mulberry wild silkworm (Bombyx mandarina) and domestic silkworm (Bombyx mori). Acta Sericologica Sinica(蚕业科学), 27, 243-252. (in Chinese with English abstract) |
| 23 | Lu C (鲁成), Yu HS (余红仕), Xiang ZH (向仲怀) (2002b) The genetic diversity and phylogenetic relationship of Bombyx mandarina and Bombyx mori from China based on RAPD analysis. Acta Entomologica Sinica(昆虫学报), 45, 198-203. (in Chinese with English abstract) |
| 24 | Moritz C, Dowling TE, Brown WM (1987) Evolution of animal mitochondrial DNA relevance for population biology and systematic. Annual Review of Ecology and Systematics, 18, 269-289. |
| 25 | Rozas J, Sánchez-DelBarrio JC, Messeguer X, Roza R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19, 2496-2497. |
| 26 | Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. |
| 27 | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| 28 | Wei S(卫斯) (1985) Primary investigation of the origin times of sericulture cultivation in China. In: Study of Agricultural History, Vol. 6(农史研究, 第六辑). China Agriculture Press, Beijing. (in Chinese) |
| 29 | Wei S (卫斯) (2005) The mark stela in our modern archeology history times: the course and importance of the first excavation in Xiyin Village historical site. . |
| 30 | Wu W (2006) Bioinformatics Analysis of the Comprise and Evolution of Animal Mitochondrial Genome (动物线粒体基因组组成与进化的生物信息学研究). PhD dissertation, Southwest University, Chongqing. (in Chinese) |
| 31 | Xia QY, Guo YR, Zhang Z, Li D, Xuan ZL, Li Z, Dai FY, Li YG, Cheng DJ, Li RQ, Cheng TC, Jiang T, Becquet C, Xu X, Liu C, Zha XF, Fan W, Lin Y, Shen YH, Jiang L, Jensen J, Hellmann I, Tang S, Zhao P, Xu HF, Yu C, Zhang GJ, Li J, Cao JJ, Liu SP, He NJ, Zhou Y, Liu H, Zhao J, Ye C, Du ZH, Pan GQ, Zhao AC, Shao HJ, Zeng W, Wu P, Li CF, Pan MH, Li JJ, Yin XY, Li DW, Wang J, Zheng HS, Wang W, Zhang XQ, Li SG, Yang HM, Lu C, Nielsen R, Zhou ZY, Wang J, Xiang ZH, Wang J (2009) Complete Resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science, 326, 433-436. |
| 32 | Xia QY (夏庆友), Zhou ZY (周泽扬), Lu C (鲁成), Xiang ZH (向仲怀) (1998) Molecular phylogenetic study on the racial differentiation of Bombyx mori by random amplified polymorphic DNA (RAPD) markers. Acta Entomologica Sinica(昆虫学报), 41, 32-40. (in Chinese with English abstract) |
| 33 | Xiang ZH(向仲怀) (1994) The Inheritance and Thremmatology of Silkworm, Bombyx mori(家蚕遗传育种学). China Agriculture Press, Beijing(in Chinese) |
| 34 | Yoshitake N (吉武成美), Jiang YL (蒋猷龙) (1987) The origin of silkworm, Bombyx mori, and its differentiation. Acta Sericologica Sinica(蚕业科学), 13, 182-184. (in Chinese) |
| 35 | Zhang H (张会), Du GS (杜桂森) (2002) The advances of molecular systematics on bryophytes. Journal of Capital Normal University (Natural Sciences Edition) (首都师范大学学报 (自然科学版) ), 23(2), 57-60. (in Chinese with English abstract) |
| 36 | Zhou KM(周匡明) (2006) The inspiration from the archeological spun silk pieces discovered at Qianshanyang site. In: Collectanea of Silk History (蚕业史论文选). Literature and History Press in China, Beijing. (in Chinese) |
| 37 | Zhou KM (周匡明) (1982) Study on the origin of sericulture cultivation. Agricultural Archaeology(农业考古), (1), 133-138. (in Chinese) |
| 38 | Zhou ZY (周泽扬), Li B (李斌), Ravikumar G, Quan GX (全国兴), Tamura T (田村俊树) (2004) Evolution studies on domestic silkworm (Bombyx mori L.) by AFLP markers. Southwest China Journal of Agricultural Sciences(西南农业学报), 17, 240-243. (in Chinese with English abstract) |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Xiaoning Zeng, Penghang Wang, Mengfan Zhang, Jing Su, Zhiyuan Shi, Fuling Gao, Jiamei Li. List of the wild woody plants in Henan Province [J]. Biodiv Sci, 2023, 31(6): 22306-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [11] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [12] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [13] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [14] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [15] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()