



Biodiv Sci ›› 2019, Vol. 27 ›› Issue (3): 314-326. DOI: 10.17520/biods.2018339 cstr: 32101.14.biods.2018339
• Original Papers • Previous Articles Next Articles
Yang Siqi1,Zhang Qi1,Song Xiqiang1,Wang Jian1,Li Yide2,Xu Han2,Guo Shouyu3,Ding Qiong1,*( )
)
Received:2018-12-25
Accepted:2019-02-27
Online:2019-03-20
Published:2019-04-09
Contact:
Ding Qiong
Yang Siqi,Zhang Qi,Song Xiqiang,Wang Jian,Li Yide,Xu Han,Guo Shouyu,Ding Qiong. Structural features of root-associated fungus-plant interaction networks in the tropical montane rain forest of Jianfengling, China[J]. Biodiv Sci, 2019, 27(3): 314-326.
| 宿主植物 Host plant | 根样品数 No. of root samples | 真菌物种数 / reads No. of fungal species/reads | 核心真菌物种数 / reads No. of core fungal species/reads | Shannon多样性指数 Shannon diversity index |
|---|---|---|---|---|
| 槭树科 Aceraceae | 34 | 701 / 31,589 | 328 / 29,817 | 3.97 |
| 番荔枝科 Annonaceae | 32 | 440 / 31,588 | 256 / 30,846 | 2.88 |
| 夹竹桃科 Apocynaceae | 41 | 434 / 31,589 | 277 / 31,134 | 2.58 |
| 冬青科 Aquifoliaceae | 39 | 675 / 31,588 | 334 / 29,932 | 4.05 |
| 棕榈科 Arecaceae | 21 | 436 / 31,589 | 264 / 30,894 | 3.12 |
| 壳斗科 Fagaceae | 31 | 359 / 31,589 | 217 / 30,983 | 2.58 |
| 樟科 Lauraceae | 35 | 463 / 31,589 | 261 / 30,508 | 4.04 |
| 木犀科 Oleaceae | 34 | 546 / 31,589 | 302 / 30,527 | 3.72 |
Table 1 Species diversity and richness of root-associated fungal community of eight plant families in the tropical montane rain forest of Jianfengling
| 宿主植物 Host plant | 根样品数 No. of root samples | 真菌物种数 / reads No. of fungal species/reads | 核心真菌物种数 / reads No. of core fungal species/reads | Shannon多样性指数 Shannon diversity index |
|---|---|---|---|---|
| 槭树科 Aceraceae | 34 | 701 / 31,589 | 328 / 29,817 | 3.97 |
| 番荔枝科 Annonaceae | 32 | 440 / 31,588 | 256 / 30,846 | 2.88 |
| 夹竹桃科 Apocynaceae | 41 | 434 / 31,589 | 277 / 31,134 | 2.58 |
| 冬青科 Aquifoliaceae | 39 | 675 / 31,588 | 334 / 29,932 | 4.05 |
| 棕榈科 Arecaceae | 21 | 436 / 31,589 | 264 / 30,894 | 3.12 |
| 壳斗科 Fagaceae | 31 | 359 / 31,589 | 217 / 30,983 | 2.58 |
| 樟科 Lauraceae | 35 | 463 / 31,589 | 261 / 30,508 | 4.04 |
| 木犀科 Oleaceae | 34 | 546 / 31,589 | 302 / 30,527 | 3.72 |
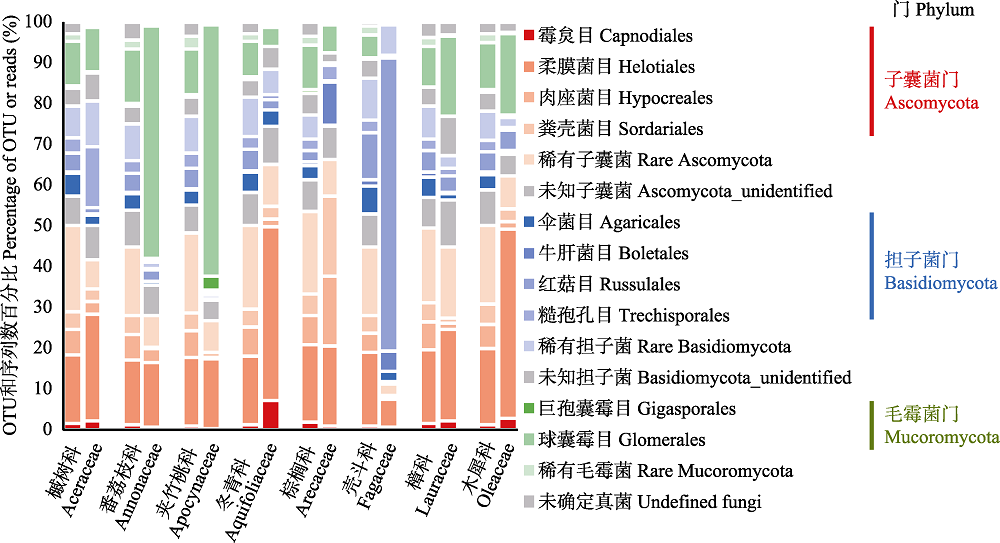
Fig. 2 Compositional differences of root-associated fungi by order among the eight plant families in the tropical montane rain forest of Jianfengling. For each plant family, left and right column are percentage of OTUs and reads, respectively.
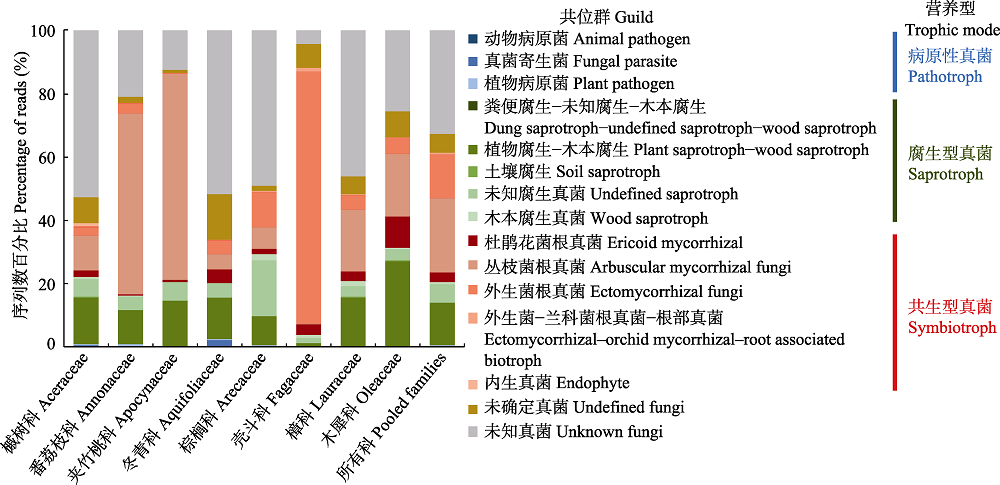
Fig. 3 Compositional differences in reads of root-associated fungi by guilds and trophic modes of the eight plant families in the tropical montane rain forest of Jianfengling
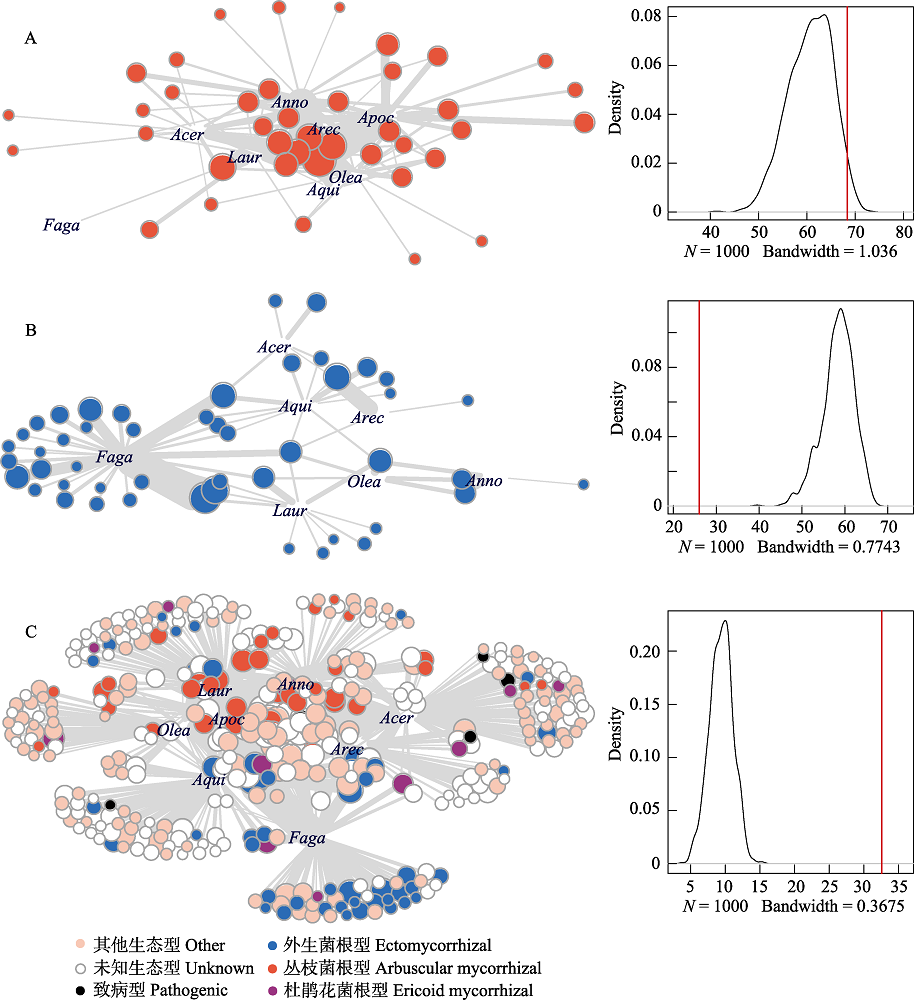
Fig. 4 Root-associated fungus-plant interaction networks (left) with density plots showing the distribution of nestedness (NODF) predicted by Patefiled’s null model (right) and observed nestedness (red vertical line) in tropical montane rain forest of Jianfenling Mountain Hainan Island. (A) and (B) are partial networks of AM and ECM interactions, and (C) is the whole root-associated fungus-plant interaction network. Acer, Aceraceae; Anno, Annoaceae; Apoc, Apocynaceae; Aqui, Aquifoliaceae; Arec, Arecaceae; Faga, Fagaceae; Laur, Lauraceae; Olea, Oleaceae. Interactions between plant and fungi are indicated by grey lines with thickness proportional to interaction strength.
| 丛枝菌根网络 Arbuscular mycorrhizal network | 外生菌根网络 Ectomycorrhizal network | 根部真菌-植物互作网络 Root-associated fungus-plant network | ||||
|---|---|---|---|---|---|---|
| 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | |
| 网络嵌套性 Nestedness metric based on overlap and decreasing fill, NODF | 68.39 | 60.70 | 26.06 | 58.36 | 32.59 | 9.40 |
| 网络加权嵌套性 Weighted NODF | 45.12 | 53.76 | 18.04 | 48.44 | 20.46 | 7.69 |
| 网络专一性 Specialization (H°2) | 0.31 | 0.01 | 0.81 | 0.01 | 0.54 | 0.01 |
| 网络连接性 Connectance | 0.42 | 0.88 | 0.21 | 0.89 | 0.26 | 0.99 |
| 植物生态位重叠 Niche overlap of plants | 0.46 | 0.99 | 0.11 | 0.99 | 0.25 | 0.99 |
| 真菌生态位重叠 Niche overlap of fungi | 0.35 | 0.97 | 0.39 | 0.98 | 0.19 | 0.91 |
| 植物的伙伴多样性 Generality of plants | 5.46 | 7.87 | 5.32 | 10.92 | 34.14 | 87.31 |
| 真菌的伙伴多样性 Vulnerability of fungi | 3.41 | 4.84 | 1.28 | 2.74 | 3.16 | 7.95 |
| 植物Checkboard值 C-score of plants | 0.17 | 0.28 | 0.62 | 0.48 | 0.38 | 0.96 |
| 真菌Checkboard值 C-score of fungi | 0.24 | 0.02 | 0.52 | 0.43 | 0.62 | 0.85 |
Table 2 Structural features of root-associated fungus-plant interaction networks
| 丛枝菌根网络 Arbuscular mycorrhizal network | 外生菌根网络 Ectomycorrhizal network | 根部真菌-植物互作网络 Root-associated fungus-plant network | ||||
|---|---|---|---|---|---|---|
| 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | |
| 网络嵌套性 Nestedness metric based on overlap and decreasing fill, NODF | 68.39 | 60.70 | 26.06 | 58.36 | 32.59 | 9.40 |
| 网络加权嵌套性 Weighted NODF | 45.12 | 53.76 | 18.04 | 48.44 | 20.46 | 7.69 |
| 网络专一性 Specialization (H°2) | 0.31 | 0.01 | 0.81 | 0.01 | 0.54 | 0.01 |
| 网络连接性 Connectance | 0.42 | 0.88 | 0.21 | 0.89 | 0.26 | 0.99 |
| 植物生态位重叠 Niche overlap of plants | 0.46 | 0.99 | 0.11 | 0.99 | 0.25 | 0.99 |
| 真菌生态位重叠 Niche overlap of fungi | 0.35 | 0.97 | 0.39 | 0.98 | 0.19 | 0.91 |
| 植物的伙伴多样性 Generality of plants | 5.46 | 7.87 | 5.32 | 10.92 | 34.14 | 87.31 |
| 真菌的伙伴多样性 Vulnerability of fungi | 3.41 | 4.84 | 1.28 | 2.74 | 3.16 | 7.95 |
| 植物Checkboard值 C-score of plants | 0.17 | 0.28 | 0.62 | 0.48 | 0.38 | 0.96 |
| 真菌Checkboard值 C-score of fungi | 0.24 | 0.02 | 0.52 | 0.43 | 0.62 | 0.85 |
| 1 |
Almario J, Jeena G, Wunder J, Langen G, Zuccaro A, Coupland G, Bucher M ( 2017) Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proceedings of the National Academy of Sciences, USA, 114, 9403-9412.
DOI URL PMID |
| 2 |
Almeida-Neto M, Ulrich W ( 2011) A straightforward computational approach for measuring nestedness using quantitative matrices. Environmental Modelling and Software, 26, 173-178.
DOI URL |
| 3 |
Bahram M, Peay KG, Tedersoo L ( 2014) Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytologist, 205, 1454-1463.
DOI URL PMID |
| 4 |
Bascompte J ( 2007) Networks in ecology. Basic and Applied Ecology, 8, 485-490.
DOI URL |
| 5 |
Bascompte J, Jordano P, Melián CJ, Olesen JM ( 2003) The nested assembly of plant-animal mutualistic networks. Proceedings of the National Academy of Sciences, USA, 100, 9383-9387.
DOI URL PMID |
| 6 |
Bastolla U, Fortuna MA, Pascual-Garcia A, Ferrera A, Luque B, Bascompte J ( 2009) The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature, 458, 1018-1020.
DOI URL |
| 7 |
Blaalid R, Davey ML, Kauserud H, Carlsen T, Halvorsen R, Hoiland K, Eidesen PB ( 2014) Arctic root-associated fungal community composition reflects environmental filtering. Molecular Ecology, 23, 649-659.
DOI URL PMID |
| 8 |
Blüthgen N, Fründ J, Vázquez DP, Menzel F ( 2008) What do interaction network metrics tell us about specialization and biological traits. Ecology, 89, 3387-3399.
DOI URL PMID |
| 9 |
Burkle LA, Marlin JC, Knight TM ( 2013) Plant-pollinator interactions over 120 years: Loss of species, co-occurrence, and function. Science, 339, 1611-1615.
DOI URL PMID |
| 10 | Cannon PF, Kirk PM ( 2007) Fungal Families of the World. CABI Bioscience, Wallingford. |
| 11 |
Cardoso EJBN, Nogueira MA, Zangaro W ( 2017) Importance of mycorrhizae in tropical soils. In: Diversity and Benefits of Microorganisms from the Tropics (eds de Azevedo J, Quecine M), Springer, Cham.
DOI URL |
| 12 |
Chagnon PL, Bradley RL, Klironomos JN ( 2012) Using ecological network theory to evaluate the causes and consequences of arbuscular mycorrhizal community structure. New Phytologist, 194, 307-312.
DOI URL |
| 13 |
Chen L, Zheng Y, Gao C, Mi XC, Ma KP, Wubet T, Guo LD ( 2017) Phylogenetic relatedness explains highly interconnected and nested symbiotic networks of woody plants and arbuscular mycorrhizal fungi in a Chinese subtropical forest. Molecular Ecology, 26, 2563-2575.
DOI URL PMID |
| 14 |
Corrales A, Henkel TW, Smith ME ( 2018) Ectomycorrhizal associations in the tropics—Biogeography, diversity patterns and ecosystem roles. New Phytologist, 220, 1076-1091.
DOI URL PMID |
| 15 |
Dickie KH, Cann C, Norman EC, Bamforth CW, Muller RE ( 2001) Foam-negative materials. Journal of the American Society of Brewing Chemists, 59, 17-23.
DOI URL |
| 16 |
Edgar RC ( 2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460-2461.
DOI URL |
| 17 |
Edgar RC ( 2013) UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10, 996.
DOI URL PMID |
| 18 |
Edgar RC ( 2018) Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences, PeerJ, 6, e4652.
DOI URL |
| 19 |
Fang JY, Li YD, Zhu B, Liu GH, Zhou GY ( 2004) Community structures and species richness in the montane rain forest of Jianfengling, Hainan Island, China. Biodiversity Science, 12, 29-43. (in Chinese with English abstract)
DOI URL |
|
[ 方精云, 李意德, 朱彪, 刘国华, 周光益 ( 2004) 海南岛尖峰岭山地雨林的群落结构、物种多样性以及在世界雨林中的地位. 生物多样性, 12, 29-43.]
DOI URL |
|
| 20 |
Fortuna MA, Stouffer DB, Olesen JM, Jordano P, Mouillot D, Krasnov BR, Poulin R, Bascompte J ( 2010) Nestedness versus modularity in ecological networks: Two sides of the same coin? Journal of Animal Ecology, 79, 811-817.
DOI URL PMID |
| 21 |
Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Hutmacher RB, Dahlberg JA, Coleman-Derr D, Lemaux PG, Taylor JW ( 2019) Strong succession in arbuscular mycorrhizal fungal communities. The ISME Journal, 13, 214-226.
DOI |
| 22 |
Gotelli NJ, McCabe DJ ( 2002) Species co-occurrence: A meta-analysis of J. M. Diamond’s assembly rules model. Ecology, 83, 2091-2096.
DOI URL |
| 23 |
Huang CW, Liao YH, Ding Q ( 2017) Two sample pooling strategies revealed different root-associated fungal diversity of Rhododendron species. Acta Microbiologica Sinica, 57, 571-581. (in Chinese with English abstract)
DOI URL |
|
[ 黄彩微, 廖映辉, 丁琼 ( 2017) 两种混合样品策略对揭示杜鹃花根部真菌多样性的影响. 微生物学报, 57, 571-581.]
DOI URL |
|
| 24 |
James A, Pitchford JW, Plank MJ ( 2012) Disentangling nestedness from models of ecological complexity. Nature, 487, 227-230.
DOI URL PMID |
| 25 | Jiang YX, Lu JP ( 1991) Tropical Forest Ecosystem of Jianfengling, Hainan Island, China. Science Press, Beijing. (in Chinese) |
| [ 蒋有绪, 卢俊培 ( 1991) 中国海南岛尖峰岭热带林生态系统. 科学出版社, 北京.] | |
| 26 |
Kivlin SN, Winston GC, Goulden ML, Treseder KK ( 2014) Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecology, 12, 14-25.
DOI URL |
| 27 |
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüβler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H ( 2013) Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology, 22, 5271-5277.
DOI URL PMID |
| 28 |
Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E ( 2009) Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences, USA, 106, 18621-18626.
DOI URL |
| 29 |
Merges D, Bálint M, Schmitt I, Böhning-Gaese K, Neuschulz EL ( 2018) Spatial patterns of pathogenic and mutualistic fungi across the elevational range of a host plant. Journal of Ecology, 106, 1545-1557.
DOI URL |
| 30 |
Newsham KK ( 2011) A meta-analysis of plant responses to dark septate root endophytes. New Phytologist, 190, 783-793.
DOI URL |
| 31 |
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG ( 2016) FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology, 20, 241-248.
DOI URL |
| 32 |
Patefield WM ( 1981) Algorithm AS 159: An efficient method of generating random R × C tables with given row and column totals. Journal of the Royal Statistical Society, Series C (Applied Statistics), 30, 91-97.
DOI URL |
| 33 |
Pianka ER ( 1974) Niche overlap and diffuse competition. Proceedings of the National Academy of Sciences, USA, 71, 2141-2145.
DOI URL PMID |
| 34 | Smith DP, Peay KG ( 2014) Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE, 9, e90234. |
| 35 | Smith SE, Read DJ ( 2008) Mycorrhizal Symbiosis, 3rd edn. Academic Press, London. |
| 36 |
Taudiere A, Munoz F, Lesne A, Monnet AC, Bellanger JM, Selosse MA, Moreau PA, Richard F ( 2015) Beyond ectomycorrhizal bipartite networks: Projected networks demonstrate contrasted patterns between early- and late-successional plants in Corsica. Frontiers in Plant Science, 6, 881.
DOI URL PMID |
| 37 | Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, Kesel AD, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K ( 2014) Global diversity and geography of soil fungi. Science, 346, 1078-1088. |
| 38 |
Thebault E, Fontaine C ( 2010) Stability of ecological communities and the architecture of mutualistic and trophic Networks. Science, 329, 853-856.
DOI URL PMID |
| 39 |
Thiéry O, Vasar M, Jairus T, Davison J, Roux C, Kivistik PA, Metspalu A, Milani L, Saks ü, Moora M, Zobel M, Öpik M ( 2016) Sequence variation in nuclear ribosomal small subunit, internal transcribed spacer and large subunit regions of Rhizophagus irregularis and Gigaspora margarita is high and isolate-dependent. Molecular Ecology, 25, 2816-2832.
DOI URL |
| 40 |
Toju H, Sato H, Yamamoto S, Kadowaki K, Tanabe AS, Yazawa S, Nishimura O, Agata K ( 2013 a) How are plant and fungal communities linked to each other in belowground ecosystems? A massively parallel pyrosequencing analysis of the association specificity of root-associated fungi and their host plants. Ecology and Evolution, 3, 3112-3124.
DOI URL PMID |
| 41 |
Toju H, Yamamoto S, Sato H, Tanabe AS, Gilbert GS, Kadowaki K ( 2013 b) Community composition of root-associated fungi in a Quercus-dominated temperate forest: “Codominance” of mycorrhizal and root-endophytic fungi. Ecology and Evolution, 3, 1281-1293.
DOI URL |
| 42 |
Tylianakis JM ( 2009) Warming up food webs. Science, 323, 1300-1301.
DOI URL |
| 43 |
Ulrich W, Gotelli NJ ( 2007) Null model analysis of species nestedness patterns. Ecology, 88, 1824-1831.
DOI URL PMID |
| 44 |
Vázquez DP, Chacoff NP, Cagnolo L ( 2009) Evaluating multiple determinants of the structure of plant-animal mutualistic networks. Ecology, 90, 2039-2046.
DOI URL PMID |
| 45 |
Xu H, Li YD, Lin MX, Wu JH, Luo TS, Zhou Z, Chen DX, Yang H, Li GJ, Liu SR ( 2015) Community characteristics of a 60 ha dynamics plot in the tropical montane rain forest in Jianfengling, Hainan Island. Biodiversity Science, 23, 192-201. (in Chinese with English abstract)
DOI URL |
|
[ 许涵, 李意德, 林明献, 吴建辉, 骆土寿, 周璋, 陈德祥, 杨怀, 李广建, 刘世荣 ( 2015) 海南尖峰岭热带山地雨林60 ha动态监测样地群落结构特征. 生物多样性, 23, 192-201.]
DOI URL |
|
| 46 |
Yamamoto S, Sato H, Tanabe AS, Hidaka A, Kadowaki K, Toju H ( 2014) Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS ONE, 9, e96363.
DOI URL PMID |
| 47 |
Zhang J, Kobert K, Flouri T, Stamatakis A ( 2013) PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics, 30, 614-620.
DOI URL PMID |
| [1] | Zhiqing Hu, Lu Dong. Effects of urbanization on interspecific interactions involving birds [J]. Biodiv Sci, 2024, 32(8): 24048-. |
| [2] | Zhirong Feng, Mingbo Chen, Xiaofang Yang, Gang Wang, Yiyi Dong, Yanqiong Peng, Huayan Chen, Bo Wang. The Ficus breeding system and the resource utilization of fig wasps shape the fig wasp community [J]. Biodiv Sci, 2024, 32(3): 23307-. |
| [3] | Peng Wang, Jiarong Sui, Xinyao Ding, Weizhong Wang, Xueqian Cao, Haipeng Zhao, Yanping Wang. Nested distribution patterns of bird assemblages and their influencing factors in Zhengzhou urban parks [J]. Biodiv Sci, 2024, 32(3): 23359-. |
| [4] | Chaoya Wang, Jintao Li, Chang Liu, Bo Wang, Baige Miao, Yanqiong Peng. Interannual stability in butterfly diversity and the larvae-plant interaction network structure at Xishuangbanna Tropical Botanical Garden [J]. Biodiv Sci, 2023, 31(12): 23305-. |
| [5] | Yanping Wang, Minchu Zhang, Chengxiu Zhan. A review on the nested distribution pattern (nestedness): Analysis methods, mechanisms and conservation implications [J]. Biodiv Sci, 2023, 31(12): 23314-. |
| [6] | Lujia Tian, Xiaobo Yang, Donghai Li, Long Li, Lin Chen, Caiqun Liang, Peichun Zhang, Chendi Li. Species diversity and nestedness of bird assemblages in the forest fragments of Haikou and Sanya cities [J]. Biodiv Sci, 2022, 30(6): 21424-. |
| [7] | Mengjun Qu, Nueryila·Ababaike , Xuge Zou, Hang Zhao, Weilin Zhu, Jianming Wang, Jingwen Li. Influence of geographic distance and environmental factors on beta diversity of plants in the Alxa gobi region in northern China [J]. Biodiv Sci, 2022, 30(11): 22029-. |
| [8] | Jiayun He, Dong Zhang, Ling Chu, Yunzhi Yan. Anthropogenic disturbances affect the functional diversity of stream fishes and its longitudinal patterns in China [J]. Biodiv Sci, 2021, 29(7): 927-937. |
| [9] | Jinfeng Zheng, Rong Tang, Shuang He, Yuehong Chen, Su Wu, Kai Zhang, Yu Xu, Xiao Zou. Bird diversity and nestedness on fragmented woodlots in Huaxi University Town, Guizhou Province [J]. Biodiv Sci, 2021, 29(5): 661-667. |
| [10] | Jian Wang, Yiyi Dong, Libin Ma, Bo Pan, Fangzhou Ma, Hui Ding, Yaping Hu, Yanqiong Peng, Xiaobing Wu, Bo Wang. Spatial variation in ant-tree network organization in the Xishuangbanna National Nature Reserve [J]. Biodiv Sci, 2020, 28(6): 695-706. |
| [11] | Mingjia Li, Kaiyuan Wu, Fanfan Meng, Ji Shen, Yongqin Liu, Nengwen Xiao, Jianjun Wang. Beta diversity of stream bacteria in Hengduan Mountains: The effects of climatic and environmental variables [J]. Biodiv Sci, 2020, 28(12): 1570-1580. |
| [12] | Dong Zhang, Fengying Wan, Ling Chu, Yunzhi Yan. Longitudinal patterns in α and β diversity of the taxonomic and functional organizations of stream fish assemblages in the Qingyi River [J]. Biodiv Sci, 2018, 26(1): 1-13. |
| [13] | Xingfeng Si, Yuhao Zhao, Chuanwu Chen, Peng Ren, Di Zeng, Lingbing Wu, Ping Ding. Beta-diversity partitioning: methods, applications and perspectives [J]. Biodiv Sci, 2017, 25(5): 464-480. |
| [14] | Xuemei Zhang, Xufang Han, Liwei Liu, Aichun Xu. Influencing factors of the nested distribution of butterfly assemblages in the Zhoushan Archipelago, China [J]. Biodiv Sci, 2016, 24(3): 321-331. |
| [15] | Qiang Fang, Shuangquan Huang. Progress in pollination networks: network structure and dynamics [J]. Biodiv Sci, 2012, 20(3): 300-307. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()