



Biodiv Sci ›› 2019, Vol. 27 ›› Issue (2): 140-148. DOI: 10.17520/biods.2018232 cstr: 32101.14.biods.2018232
• Original Papers • Previous Articles Next Articles
Fan Jingyu,Li Hanpeng,Yang Zhuo,Zhu Gengping
Received:2018-08-31
Accepted:2018-12-08
Online:2019-02-20
Published:2019-04-16
Contact:
Zhu Gengping
Fan Jingyu, Li Hanpeng, Yang Zhuo, Zhu Gengping. Selecting the best native individual model to predict potential distribution of Cabomba caroliniana in China[J]. Biodiv Sci, 2019, 27(2): 140-148.
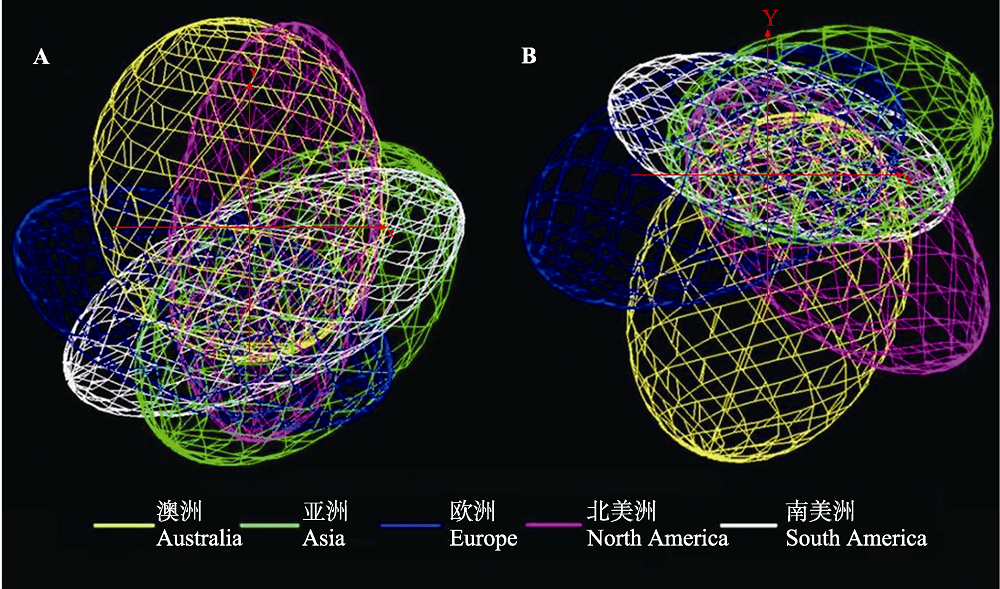
Fig. 2 Comparison of climate niche spaces occupied by different populations in five continent. Left: A dataset, including Bio2, Bio3, Bio10, Bio11, Bio15, Bio16, Bio18, and Bio19; Right: B dataset, including Bio1, Bio5, Bio6, Bio12, Bio13, Bio14, and Bio15.
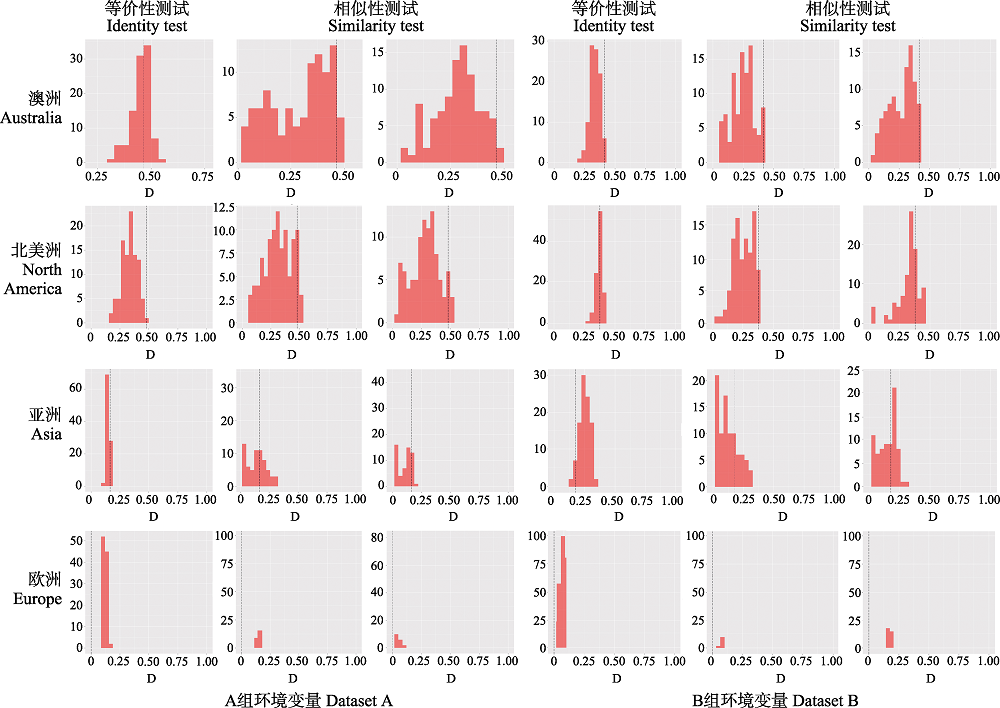
Fig. 3 Histograms of niche equivalency and similarity tests between continental populations based on the two environmental datasets. Black dotted lines represent the observed niche overlap. whereas black bars represent simulated niche overlaps. Left: A dataset; Right: B dataset. Details of dataset A and B refer to Fig. 2.
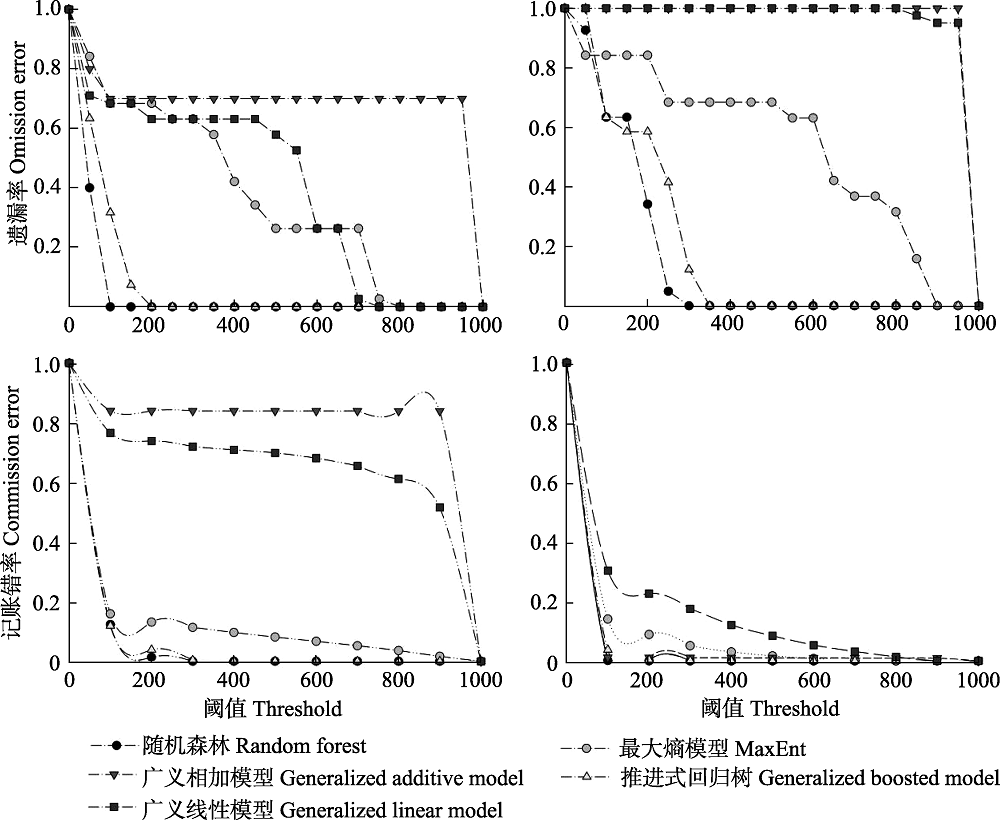
Fig. 4 Omission and commission error comparisons of native models transferability in China based on the two datasets. Left: A dataset; Right: B dataset. Details of dataset A and B refer to Fig. 2.
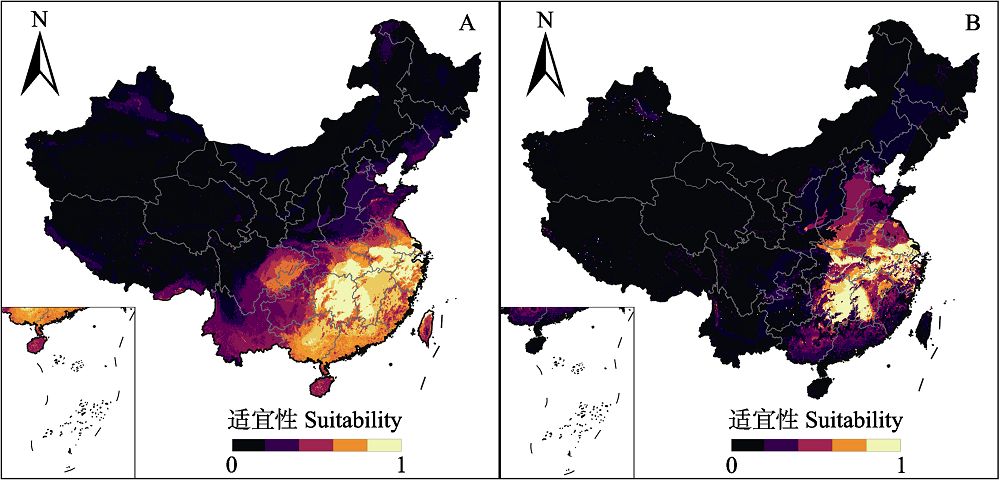
Fig. 5 Potential distributions of Cabomba caroliniana based on the two native random forest models using two environmental datasets. Left: A dataset; Right: B dataset. Details of dataset A and B refer to Fig. 2.
| [7] |
[ 丁炳扬, 于明坚, 金孝锋, 余建, 姜维梅, 董柯锋 ( 2003) 水盾草在中国的分布特点和入侵途径. 生物多样性, 11, 223-230.]
DOI URL |
| [8] |
Dong ZD, Shi LC, Huang GZ, Qin YC ( 2007) Investigation and countermeasure of plant invasion in Liuzhou. Guangxi Plant Protection, 20(2), 27-29. (in Chinese)
DOI URL |
|
[ 董志德, 石亮成, 黄桂珍, 覃燕城 ( 2007) 柳州市生态灾害植物外侵种的调查与对策. 广西植保, 20(2), 27-29.]
DOI URL |
|
| [9] |
Evans SN, Shvets Y, Slatkin M ( 2007) Non-equilibrium theory of the allele frequency spectrum. Theoretical Population Biology, 71, 109-119.
DOI URL |
| [10] |
Fan JY, Zhao NX, Li M, Wang ML, Zhu GP ( 2018) What are the best predictors for invasive potential of weeds? Transferability evaluations of model predictions based on diverse environmental data sets for Flaveria bidentis. Weed Research, 58, 141-149.
DOI URL |
| [11] |
He JX, Huang C, Wan FH, Guo JY ( 2011) Invasion and distribution of Cabomba caroliniana in wetlands of Jiangsu, China. Chinese Journal of Applied and Environmental Biology, 17, 186-190. (in Chinese with English abstract)
DOI URL |
|
[ 何金星, 黄成, 万方浩, 郭建英 ( 2011) 水盾草在江苏省重要湿地的入侵与分布现状. 应用与环境生物学报, 17, 186-190.]
DOI URL |
|
| [12] |
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A ( 2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978.
DOI URL |
| [13] |
Hou YT, Shu FY, Dong SX, Liu M, Xie SG ( 2012) Cabomba caroliniana, a new recorded exotic aquatic plant in Nansi Lake and its habitat characters. Acta Hydrobiologica Sinica, 36, 1005-1008. (in Chinese with English abstract)
DOI URL |
|
[ 侯元同, 舒凤月, 董士香, 柳明, 谢松光 ( 2012) 水盾草——南四湖外来水生植物新记录及其生境特点. 水生生物学报, 36, 1005-1008.]
DOI URL |
|
| [14] |
Peterson AT, Papeş M, Soberón J ( 2008) Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecological Modelling, 213, 63-72.
DOI URL |
| [15] |
Peterson AT, Cobos ME, Jiménez-García D ( 2018) Major challenges for correlational ecological niche model projections to future climate conditions. Annals of the New York Academy of Sciences, 1429, 66-77.
DOI URL PMID |
| [16] |
Peterson AT, Soberón J ( 2012) Species distribution modeling and ecological niche modeling: Getting the concepts right. Brazilian Journal for Nature Conservation, 10, 1-6.
DOI URL |
| [17] |
Petitpierre B, Kueffer C, Broennimann O, Randin C, Daehler C, Guisan A ( 2012) Climatic niche shifts are rare among terrestrial plant invaders. Science, 335, 1344-1348.
DOI URL PMID |
| [18] |
Qiao HJ, Peterson AT, Campbell LP, Soberón J, Ji LQ, Escobar LE ( 2016) NicheA: Creating virtual species and ecological niches in multivariate environmental scenarios. Ecography, 39, 805-813.
DOI URL |
| [19] | Qiao HJ, Feng X, Escobar LE, Peterson AT, Soberón J, Zhu GP, Papeş M ( 2018) An evaluation of transferability of ecological niche models. Ecography, 30, 550-560. |
| [20] | Shen ZH ( 2000) Cabomba caroliniana—New invasive alien species. Life World, ( 2), 37-38. (in Chinese) |
| [ 沈脂红 ( 2000) 水盾草——新入侵的外来种. 生命世界, ( 2), 37-38.] | |
| [21] | Soberón J , Nakamura ( 2009) Niches and distributional areas: Concepts, methods and assumptions. Proceedings of the National Academy of Sciences, USA, 106, 19644-19650. |
| [22] |
Thuiller W ( 2003) BIOMOD—Optimizing predictions of species distributions and projecting potential future shifts under global change. Global Change Biology, 9, 1353-1362.
DOI URL |
| [23] |
Tingley R, Meiri S, Chapple DG ( 2016) Addressing knowledge gaps in reptile conservation. Biological Conservation, 204, 1-5.
DOI URL |
| [24] |
Uden DR, Allen CR, Angeler DG, Corral LL, Fricke K ( 2015) Adaptive invasive species distribution models: A framework for modeling incipient invasions. Biological Invasions, 17, 2831-2850.
DOI URL |
| [25] |
Wan ZG, Gu YJ, Qian SX ( 1999) Cabomba Aubl. of Nymphaeaceae, a new record genus from China. Journal of Wuhan Botanical Research, 17, 215-216. (in Chinese with English abstract)
DOI URL |
|
[ 万志刚, 顾咏洁, 钱士心 ( 1999) 中国睡莲科一新记录属——水盾草属. 武汉植物学研究, 17, 215-216.]
DOI URL |
|
| [26] | Warren DL, Beaumont L, Dinnage R, Baumgartner J ( 2018) New methods for measuring ENM breadth and overlap in environmental space. Ecography, 42, 444-446. |
| [27] |
Xu CY, Zhang WJ, Lu BR, Chen JK ( 2001) Progress in studies on mechanisms of biological invasion. Biodiversity Science, 9, 430-438. (in Chinese with English abstract)
DOI URL |
| [1] | Bai YZ, Cao XF, Chen C, Hu BS, Liu FQ ( 2009) Potential distribution areas of alien invasive plant Flaveria bidentis (Asteraceae) in China. Chinese Journal of Applied Ecology, 20, 2377-2383. (in Chinese with English abstract) |
| [ 白艺珍, 曹向锋, 陈晨, 胡白石, 刘凤权 ( 2009) 黄顶菊在中国的潜在适生区. 应用生态学报, 20, 2377-2383.] | |
| [27] |
[ 徐承远, 张文驹, 卢宝荣, 陈家宽 ( 2001) 生物入侵机制研究进展. 生物多样性, 9, 430-438.]
DOI URL |
| [28] |
Yates KL, Bouchet PJ, Caley M, Mengersen K, Randin CF, Parnell S, Fielding AH, Bamford AJ, Ban S, Barbosa AM, Dormann CF, Elith J, Embling CB, Ervin GN, Fisher R, Gould S, Graf RF, Gregr EJ, Sequeira AMM ( 2018) Outstanding challenges in the transferability of Ecological Models. Trends in Ecology and Evolution, 33, 790-802.
DOI URL |
| [2] |
Breiner FT, Guisan A, Bergamini A, Nobis MP ( 2015) Overcoming limitations of modelling rare species by using ensembles of small models. Methods in Ecology and Evolution, 6, 1210-1218.
DOI URL |
| [3] |
Brown JL ( 2014) SDMtoolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods in Ecology and Evolution, 5, 694-700.
DOI URL |
| [29] | Zhang JL, Meng SY ( 2013) Cabomba caroliniana A. Gray found in Beijing water areas. Weed Science, 31(2), 45-46. (in Chinese) |
| [ 张劲林, 孟世勇 ( 2013) 北京水域发现水盾草. 杂草科学, 31(2), 45-46.] | |
| [4] | Ding BY ( 1999) The first occurrence of Cabomba caroliniana A. Gray (Cabombaceae) in China. Journal of Hangzhou University (Nature Science), 26, 97. (in Chinese) |
| [ 丁炳扬 ( 1999) 绿菊花草在我国首次发现. 杭州大学学报(自然科学版), 26, 97.] | |
| [30] | Zhang Q ( 2012) The aquatic plant of Cabomba caroliniana. Garden, ( 8), 74-75. (in Chinese) |
| [ 张群 ( 2012) 水生植物水盾草. 园林, ( 8), 74-75.] | |
| [31] |
Zhu GP, Fan JY, Wang ML, Chen M, Qiao HJ ( 2017) The importance of the shape of receiver operating characteristic (ROC) curve in ecological model evaluation—Case study of Hlyphantria cunea. Journal of Biosafety, 26, 184-190. (in Chinese with English abstract)
DOI URL |
|
[ 朱耿平, 范靖宇, 王梦琳, 陈敏, 乔慧捷 ( 2017) ROC曲线形状在生态位模型评价中的重要性——以美国白蛾为例. 生物安全学报, 26, 184-190.]
DOI URL |
|
| [5] |
Ding BY ( 2000) Cabomba Aublet (Cabombaceae), a newly naturalized genus of China. Acta Phytotaxonomica Sinica, 38, 198-200. (in Chinese with English abstract)
DOI URL |
|
[ 丁炳扬 ( 2000) 中国水生植物一新归化属——水盾草属(莼菜科). 植物分类学报, 38, 198-200.]
DOI URL |
|
| [32] |
Zhu GP, Gariepy TD, Haye T, Bu WJ ( 2017) Patterns of niche filling and expansion across the invaded ranges of Halyomorpha halys in North America and Europe. Journal of Pest Science, 90, 1045-1057.
DOI URL |
| [33] | Zhu GP, Liu GQ, Bu WJ, Gao YB ( 2013) Ecological niche modeling and its applications in biodiversity conservation. Biodiversity Science, 21, 90-98. (in Chinese with English abstract) |
| [ 朱耿平, 刘国卿, 卜文俊, 高玉葆 ( 2013) 生态位模型的基本原理及其在生物多样性保护中的应用. 生物多样性, 21, 90-98.] | |
| [34] |
Zhu GP, Liu Q, Gao YB ( 2014) Improving ecological niche model transferability to predict the potential distribution of invasive exotic species. Biodiversity Science, 22, 223-230. (in Chinese with English abstract)
DOI URL |
| [6] | Ding BY, Jin XF, Yu MJ, Yu J, Shen HM, Wang YF ( 2007) Impact to native species by invaded subaqueous plant Cabomba caroliniana. Oceanologia et Limnologia Sinica, 38, 336-342. (in Chinese with English abstract) |
| [ 丁炳扬, 金孝锋, 于明坚, 余建, 沈海铭, 王月丰 ( 2007) 水盾草(Cabomba caroliniana)入侵对沉水植物群落物种多样性组成的影响. 海洋与湖沼, 38, 336-342.] | |
| [34] |
[ 朱耿平, 刘强, 高玉葆 ( 2014) 提高生态位模型转移能力来模拟入侵物种的潜在分布. 生物多样性, 22, 223-230.]
DOI URL |
| [35] |
Zhu GP, Peterson T ( 2017) Do consensus models outperform individual models? Transferability evaluations of diverse modeling approaches for an invasive moth. Biological Invasions, 19, 2519-2532.
DOI URL |
| [7] |
Ding BY, Yu MJ, Jin XF, Yu J, Jiang WM, Dong KF ( 2003) The distribution characteristics and invasive route of Cabomba caroliniana in China. Biodiversity Science, 11, 223-230. (in Chinese with English abstract)
DOI URL |
| [36] |
Živković D, Stephan W ( 2011) Analytical results on the neutral non-equilibrium allele frequency spectrum based on diffu?sion theory. Theoretical Population Biology, 79, 184-191.
DOI URL PMID |
| [1] | Zhou Zhihua, Jin Xiaohua, Luo Ying, Li Diqiang, Yue Jianbing, Liu Fang, He Tuo, Li Xi, Dong Hui, Luo Peng. Analyses and suggestions on mechanisms of forestry and grassland administrations in China to achieve targets of Kunming-Montreal Global Biodiversity Framework [J]. Biodiv Sci, 2025, 33(3): 24487-. |
| [2] | Tian Zhiqi, Su Yang. China’s implementation model of international environmental conventions and its application to the Convention on Biological Diversity—Achieving the Kunming-Montreal Global Biodiversity Framework targets and the role of national parks [J]. Biodiv Sci, 2025, 33(3): 24593-. |
| [3] | Wei Yi, Yi Ai, Meng Wu, Liming Tian, Tserang Donko Mipam. Soil archaeal community responses to different grazing intensities in the alpine meadows of the Qinghai-Tibetan Plateau [J]. Biodiv Sci, 2025, 33(1): 24339-. |
| [4] | Bangze Li, Shuren Zhang. An updated species checklist and taxonomic synopsis of Cyperaceae in China [J]. Biodiv Sci, 2024, 32(7): 24106-. |
| [5] | Zonggang Hu. China and the United States once planned joint compilation of the Flora of China after the victory in the China’s Resistance War Against Japan [J]. Biodiv Sci, 2024, 32(6): 24220-. |
| [6] | Qiyu Kuang, Liang Hu. Species diversity and geographical distribution of marine benthic shell-bearing mollusks around Donghai Island and Naozhou Island, Guangdong Province [J]. Biodiv Sci, 2024, 32(5): 24065-. |
| [7] | Yujie Chi, Xintian Zhang, Zhixuan Tian, Chengshuai Guan, Xinzhi Gu, Zhihui Liu, Zhanbin Wang, Jinjie Wang. Species diversity of powdery mildew fungi (Erysiphaceae) and their host plants in Northeast Asia [J]. Biodiv Sci, 2024, 32(4): 23443-. |
| [8] | Di Lin, Shuanglin Chen, Que Du, Wenlong Song, Gu Rao, Shuzhen Yan. Investigation of species diversity of myxomycetes in Dabie Mountains [J]. Biodiv Sci, 2024, 32(2): 23242-. |
| [9] | Jianping Jiang, Bo Cai, Bin Wang, Weitao Chen, Zhixin Wen, Dezhi Zhang, Lulu Sui, Shun Ma, Weibo Wang. New vertebrate species discovered in China in 2023 [J]. Biodiv Sci, 2024, 32(11): 24327-. |
| [10] | Huanxi Cao, Qingsong Zhou, Arong Luo, Pu Tang, Tingjing Li, Zejian Li, Huayan Chen, Zeqing Niu, Chaodong Zhu. New taxa of extant Hymenoptera in 2023 [J]. Biodiv Sci, 2024, 32(11): 24319-. |
| [11] | Cheng Du, Jun Liu, Wen Ye, Shuai Liao. 2023 annual report on new taxa and nomenclatural changes for Chinese plants [J]. Biodiv Sci, 2024, 32(11): 24253-. |
| [12] | Siyuan Xu, Qiqi Lian, Ruixin Zhang, Jiateng Zhao, Xuan Zhou, Lu Zhou, Qin Chen, Ming Bai. The world new taxa of extant Coleoptera in 2023 [J]. Biodiv Sci, 2024, 32(11): 24307-. |
| [13] | Nuriye Muhetaier, Xiuying Zhang, Subinuer Eli, Houhun Li. Annual report of new taxa for Chinese Lepidoptera in 2023 [J]. Biodiv Sci, 2024, 32(11): 24428-. |
| [14] | Chen Lin, Qicheng Yang, Yanling Wu, Peng Hou, Bing Zhang, Ding Yang. New taxa of Diptera from China in 2023 [J]. Biodiv Sci, 2024, 32(11): 24328-. |
| [15] | Yuchen Du, Beimeng Liu, Junfeng Chen, Hao Wang, Yi Xie. Analysis of factors influencing farmers’ protection willingness based on structural equation model: Taking the Hunchun area of Northeast China Tiger and Leopard National Park as an example [J]. Biodiv Sci, 2024, 32(1): 23155-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()