



Biodiv Sci ›› 2015, Vol. 23 ›› Issue (5): 649-657. DOI: 10.17520/biods.2015032 cstr: 32101.14.biods.2015032
Special Issue: 森林动态监测样地专题; 土壤生物与土壤健康
• Special Feature: Forest Dynamics Monitoring • Previous Articles Next Articles
Aihua Zhao1,2, Xiaojun Du1,*( ), Jing Zang1, Shouren Zhang1, Zhihua Jiao3
), Jing Zang1, Shouren Zhang1, Zhihua Jiao3
Received:2015-02-10
Accepted:2015-02-10
Online:2015-09-20
Published:2015-10-12
Contact:
Du Xiaojun
Aihua Zhao, Xiaojun Du, Jing Zang, Shouren Zhang, Zhihua Jiao. Soil bacterial diversity in the Baotianman deciduous broad-leaved forest[J]. Biodiv Sci, 2015, 23(5): 649-657.
| 样品编号 Sample identity | 原始序列数 Original sequence number | 原始OTU数 Original OTU number | 标准化序列数 Even sequence number | 标准化后OTU数 Even OTU number |
|---|---|---|---|---|
| 0101 | 37,569 | 7,148 | 36,000 | 6,979 |
| 0105 | 39,949 | 5,671 | 36,000 | 5,320 |
| 0203 | 39,698 | 5,661 | 36,000 | 5,327 |
| 0218 | 39,337 | 5,579 | 36,000 | 5,320 |
| 0306 | 39,281 | 6,396 | 36,000 | 6,097 |
| 0318 | 39,760 | 6,263 | 36,000 | 5,906 |
| 0411 | 39,756 | 6,008 | 36,000 | 5,672 |
| 0606 | 39,636 | 6,642 | 36,000 | 6,286 |
| 0701 | 39,945 | 6,059 | 36,000 | 5,694 |
| 0819 | 39,864 | 6,324 | 36,000 | 5,933 |
| 0913 | 39,827 | 5,275 | 36,000 | 4,971 |
| 0919 | 39,720 | 5,958 | 36,000 | 5,645 |
| 1002 | 39,844 | 5,486 | 36,000 | 5,175 |
| 1006 | 39,551 | 6,658 | 36,000 | 6,322 |
| 1018 | 39,711 | 5,526 | 36,000 | 5,184 |
| 1104 | 39,746 | 6,380 | 36,000 | 6,036 |
| 1210 | 39,569 | 6,255 | 36,000 | 5,933 |
| 1212 | 39,659 | 6,016 | 36,000 | 5,691 |
| 1318 | 39,809 | 6,706 | 36,000 | 6,326 |
| 1405 | 39,825 | 5,910 | 36,000 | 5,547 |
| 1510 | 39,379 | 5,650 | 36,000 | 5,378 |
| 1512 | 37,588 | 5,555 | 36,000 | 5,413 |
| 1607 | 39,739 | 6,338 | 36,000 | 5,981 |
| 1615 | 39,212 | 6,246 | 36,000 | 5,978 |
| 1618 | 39,578 | 6,552 | 36,000 | 6,200 |
| 1704 | 39,495 | 6,405 | 36,000 | 6,024 |
| 1718 | 39,130 | 6,910 | 36,000 | 6,564 |
| 1808 | 39,739 | 6,833 | 36,000 | 6,498 |
| 1814 | 39,866 | 6,850 | 36,000 | 6,501 |
| 1902 | 39,155 | 7,283 | 36,000 | 6,920 |
| 1913 | 38,882 | 7,145 | 36,000 | 6,844 |
Table 1 Sequence numbers and OTU numbers of individual samples
| 样品编号 Sample identity | 原始序列数 Original sequence number | 原始OTU数 Original OTU number | 标准化序列数 Even sequence number | 标准化后OTU数 Even OTU number |
|---|---|---|---|---|
| 0101 | 37,569 | 7,148 | 36,000 | 6,979 |
| 0105 | 39,949 | 5,671 | 36,000 | 5,320 |
| 0203 | 39,698 | 5,661 | 36,000 | 5,327 |
| 0218 | 39,337 | 5,579 | 36,000 | 5,320 |
| 0306 | 39,281 | 6,396 | 36,000 | 6,097 |
| 0318 | 39,760 | 6,263 | 36,000 | 5,906 |
| 0411 | 39,756 | 6,008 | 36,000 | 5,672 |
| 0606 | 39,636 | 6,642 | 36,000 | 6,286 |
| 0701 | 39,945 | 6,059 | 36,000 | 5,694 |
| 0819 | 39,864 | 6,324 | 36,000 | 5,933 |
| 0913 | 39,827 | 5,275 | 36,000 | 4,971 |
| 0919 | 39,720 | 5,958 | 36,000 | 5,645 |
| 1002 | 39,844 | 5,486 | 36,000 | 5,175 |
| 1006 | 39,551 | 6,658 | 36,000 | 6,322 |
| 1018 | 39,711 | 5,526 | 36,000 | 5,184 |
| 1104 | 39,746 | 6,380 | 36,000 | 6,036 |
| 1210 | 39,569 | 6,255 | 36,000 | 5,933 |
| 1212 | 39,659 | 6,016 | 36,000 | 5,691 |
| 1318 | 39,809 | 6,706 | 36,000 | 6,326 |
| 1405 | 39,825 | 5,910 | 36,000 | 5,547 |
| 1510 | 39,379 | 5,650 | 36,000 | 5,378 |
| 1512 | 37,588 | 5,555 | 36,000 | 5,413 |
| 1607 | 39,739 | 6,338 | 36,000 | 5,981 |
| 1615 | 39,212 | 6,246 | 36,000 | 5,978 |
| 1618 | 39,578 | 6,552 | 36,000 | 6,200 |
| 1704 | 39,495 | 6,405 | 36,000 | 6,024 |
| 1718 | 39,130 | 6,910 | 36,000 | 6,564 |
| 1808 | 39,739 | 6,833 | 36,000 | 6,498 |
| 1814 | 39,866 | 6,850 | 36,000 | 6,501 |
| 1902 | 39,155 | 7,283 | 36,000 | 6,920 |
| 1913 | 38,882 | 7,145 | 36,000 | 6,844 |
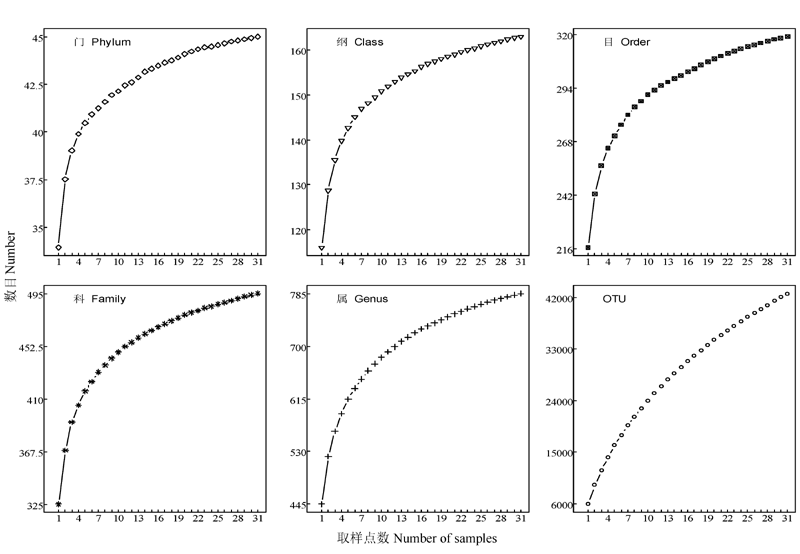
Fig. 3 Rarefaction curves of the observed bacterial quantity at different taxonomic levels varying with number of samples in the Baotianman Forest Dynamics Plot
| [1] | Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2010) Examining the global distribution of dominant archaeal populations in soil.The International Society for Microbial Ecology, 5, 908-917. |
| [2] | Cai CQ (蔡晨秋), Tang L (唐丽), Long CL (龙春林) (2011) Soil microbial diversity and its research methods.Journal of Anhui Agricultural Science(安徽农业科学), 39, 17274-17276, 17278. (in Chinese with English abstract) |
| [3] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data.Nature Methods, 7, 335-336. |
| [4] | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences,USA, 108(S1), 4516-4522. |
| [5] | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina Hiseq and Miseq platforms.The International Society for Microbial Ecology, 6, 1621-1624. |
| [6] | Chakravorty S, Helb D, Burday M, Connell N, Allan D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of Microbiological Methods, 69, 330-339. |
| [7] | Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis.Nucleic Acids Research, 42(Datbase issue), D633-D642. |
| [8] | Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM, Pelletier E, Hirsch PR, Meyer F, Gilbert JA, Le Paslier D, Simonet P, Vogel TM (2012) Structure, fluctuation and magnitude of a natural grassland soil metagenome. The International Society for Microbial Ecology, 6, 1677-1687. |
| [9] | Fernando WJD, Li R (2012) Opening the black box: understanding the influence of cropping systems and plant communities on bacterial and fungal population dynamics. Ceylon Journal of Science (Biology Science), 41, 89-110. |
| [10] | Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities.Proceedings of the National Academy of Sciences, USA, 103, 626-631. |
| [11] | Fierer N, Leff JW, Adams BJ, Nielsend UN, Thomas BS, Lauber CL, Sarah O, Gilberte JA, Wall DH, Caporaso JG (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes.Proceedings of the National Academy of Sciences, USA, 109, 21390-21395. |
| [12] | Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils.Environmental Microbiology, 13, 1642-1654. |
| [13] | Gu FX (顾峰雪), Wen QK (文启凯), Pan BR (潘伯荣), Yang YS (杨玉锁) (2000) A preliminary study on soil microorganisms of artificial vegetation in the center of Taklimakan Desert.Biodiversity Science(生物多样性), 8, 297-303. (in Chinese with English abstract) |
| [14] | Guo LD (郭良栋) (2012) Progress of microbial species diversity research in China.Biodiversity Science(生物多样性), 20, 572-580. (in Chinese with English abstract) |
| [15] | Hackl E, Pfeffer M, Donat C, Bachmann G, Zechmeister-Boltenstern S (2005) Composition of the microbial communities in the mineral soil under different types of natural forest.Soil Biology and Biochemistry, 37, 661-671. |
| [16] | Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A (2004) Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils.Applied Environmental Microbiology, 70, 5057-5065. |
| [17] | He ZL, van Nostrand JD, Deng Y, Zhou JZ (2011) Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities.Frontiers of Environmental Science and Engineering in China, 5, 1-20. |
| [18] | Jiang HY (姜海燕), Yan W (闫伟), Li XT (李晓彤), Yang XL (杨秀丽), Lv HL (吕洪丽) (2010) The diversity of soil microorganism under different vegetations of Larix gmelinii forest in Great Xing’an Mountains.Microbiology China(微生物学通报), 37, 186-190. (in Chinese with English abstract) |
| [19] | Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses.The International Society for Microbial Ecology, 3, 442-453. |
| [20] | Lauber CL, Micah H, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale.Applied and Environmental Microbiology, 75, 5111-5120. |
| [21] | Li H, Ye D, Wang X, Settles ML, Wang J, Hao Z, Zhou L, Dong P, Jiang Y, Ma ZS (Sam) (2014) Soil bacterial communities of different natural forest types in Northeast China. Plant Soil, 383, 203-216. |
| [22] | Liu YD (刘雨迪), Chen XY (陈小云), Liu MQ (刘满强), Qin JT (秦江涛), Li HX (李辉信), Hu F (胡锋) (2013) Changes in soil microbial properties and nematode assemblage over time during rice cultivation. Biodiversity Science(生物多样性), 21, 334-342. (in Chinese with English abstract) |
| [23] | Macrae A (2000) The use of 16S rDNA methods in soil microbial ecology.Brazilian Journal of Microbiology, 31, 77-82. |
| [24] | Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies.Bioinformatics, 27, 2957-2963. |
| [25] | Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils.PLoS ONE, 6, e17000. |
| [26] | Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity.The International Society for Microbial Ecology, 1, 283-290. |
| [27] | Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. The International Society for Microbial Ecology, 4, 1340-1351. |
| [28] | Shen CC, Xiong JB, Zhang HY, Feng YZ, Lin XG, Li XY, Liang WJ, Chu HY (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain.Soil Biology and Biochemistry, 57, 204-211. |
| [29] | Shi P (时鹏), Gao Q (高强), Wang SP (王淑平), Zhang Y (张妍) (2010) Effects of continuous cropping of corn and fertilization on soil microbial community functional diversity.Acta Ecologica Sinica(生态学报), 30, 6173-6182. (in Chinese with English abstract) |
| [30] | Song CS (宋朝枢) (1994) Scientific Investigation in the Baotianman Nature Reserve (宝天曼自然保护区科学考察集). China Forestry Publishing House, Beijing. (in Chinese) |
| [31] | Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology.International Journal of Systematic Bacteriology, 44, 846-849. |
| [32] | Sun X (孙欣), Gao Y (高莹), Yang YF (杨云锋) (2013) Recent advancement in microbial environmental research using metagenomics tools.Biodiversity Science(生物多样性), 21, 393-400. (in Chinese with English abstract) |
| [33] | Teng QH (滕齐辉), Cao H (曹慧), Cui ZL (崔中利), Wang Y (王英), Sun B (孙波), Hao HT (郝红涛), Li SP (李顺鹏) (2006) PCR-RFLP analysis of bacterial 16S rDNA from a typical garden soil in Taihu region.Biodiversity Science(生物多样性), 14, 345-351. (in Chinese with English abstract) |
| [34] | Tiedje JM, Asuming-Brempong S, Nüsslein K, Marsh TL, Flynn SJ (1999) Opening the black box of soil microbial diversity.Applied Soil Ecology, 13, 109-122. |
| [35] | van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems.Ecology Letters, 11, 296-310. |
| [36] | Wooley JC, Godzik A, Friedberg I (2010) A primer on metagenomics.PLoS Computational Biology, 6, e1000667. |
| [37] | Wu ZY (吴则焰), Lin WX (林文雄), Chen ZF (陈志芳), Fang CX (方长旬), Zhang ZX (张志兴), Wu LK (吴林坤), Zhou MM (周明明), Chen T (陈婷) (2013) Variations of soil microbial community diversity along an elevational gradient in mid-subtropical forest.Chinese Journal of Plant Ecology(植物生态学报), 37, 397-406. (in Chinese with English abstract) |
| [38] | Yang GP (杨官品), Nan L (男兰), Jia HB (贾海波), Zhu YH (朱艳红), Liu YJ (刘英杰), Zhang K (张凯) (2000) Bacterial genetic diversity in soils and their correlation with vegetation.Acta Genetica Sinica(遗传学报), 27, 278-282. (in Chinese with English abstract) |
| [39] | Yuan YL, Si GC, Wang J, Luo TX, Zhang GX (2014) Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau.FEMS Microbiology Ecology, 87, 121-132. |
| [40] | Zhang JP (张建萍), Dong NY (董乃源), Yu HB (余浩滨), Zhou YJ (周勇军), Lu YL (陆永良), Geng RM (耿锐梅), Yu LQ (余柳青) (2008) Bacteria diversity in paddy field soil by 16S rDNA-RFLP analysis in Ningxia.Biodiversity Science(生物多样性), 16, 586-592. (in Chinese with English abstract) |
| [41] | Zhang YG, Cong J, Lu H, Li GL, Qu YY, Su XJ, Zhou JZ, Li DQ (2014a) Community structure and elevational diversity patterns of soil Acidobacteria.Journal of Environmental Sciences, 26, 1717-1724. |
| [42] | Zhang YG, Cong J, Lu H, Yang CY, Yang YF, Zhou JZ, Li DQ (2014b) An integrated study to analyze soil microbial community structure and metabolic potential in two forest types.PLoS ONE, 9, e93773. |
| [43] | Zhao XL (赵先丽), Zhou GS (周广胜), Zhou L (周莉), Lv GH (吕国红), Jia QY (贾庆宇), Xie YB (谢艳兵) (2007) A preliminary study on soil microorganism in Panjin reed wetland.Journal of Meteorology and Environment(气象与环境学报), 23, 30-33. (in Chinese with English abstract) |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()