



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (1): 24241. DOI: 10.17520/biods.2024241 cstr: 32101.14.biods.2024241
• Original Papers: Genetic Diversity • Previous Articles Next Articles
Hong Deng1,2, Zhanyou Zhong1,2, Chunni Kou2, Shuli Zhu2, Yuefei Li2, Yuguo Xia2, Zhi Wu2, Jie Li2,*( ), Weitao Chen2,*(
), Weitao Chen2,*( )(
)( )
)
Received:2024-06-17
Accepted:2024-08-06
Online:2025-01-20
Published:2024-09-20
Contact:
* E-mail: Supported by:Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes[J]. Biodiv Sci, 2025, 33(1): 24241.
| 河流 River | 采集地 Sampling sites | 样本量 Sample size | 经纬度 Locality |
|---|---|---|---|
| 南盘江 Nanpanjiang (NPJ) | 西林 Xiling (XL) | 7 | 24˚39′ N/104˚28′ E |
| 柳江 Liujiang (LJ) | 宜州 Yizhou (YZ) | 8 | 24˚50′ N/108˚60′ E |
| 柳江 Liujiang (LJ) | 柳州 Liuzhou (LZZ) | 2 | 24˚31′ N/109˚42′ E |
| 柳江 Liujiang (LJ) | 柳城 Liucheng (LC) | 8 | 24˚65′ N/109˚26′ E |
| 桂江 Guijiang (GJ) | 昭平 Zhaoping (ZP) | 2 | 24˚17′ N/110˚80′ E |
| 桂江 Guijiang (GJ) | 平乐 Pingle (PL) | 4 | 24˚62′ N/110˚62′ E |
| 东江 Dongjiang (DJ) | 古竹 Guzhu (GZ) | 4 | 23˚51′ N/114˚70′ E |
| 东江 Dongjiang (DJ) | 惠州 Huizhou (HZ) | 2 | 23˚11′ N/114˚41′ E |
| 贺江 Hejiang (HEJ) | 南丰 Nanfeng (NF) | 5 | 23˚44′ N/111˚47′ E |
| 北江 Beijiang (BJ) | 韶关 Shaoguan (SG) | 4 | 24˚81′ N/113˚59′ E |
| 北江 Beijiang (BJ) | 连州 Lianzhou (LZ) | 8 | 24˚78′ N/112˚37′ E |
| 北江 Beijiang (BJ) | 西牛 Xiniu (XN) | 14 | 24˚17′ N/113˚11′ E |
| 红水河 Hongshuihe (HSH) | 都安 Du’an (DA) | 5 | 23˚93′ N/108˚10′ E |
| 郁江 Yujiang (YJ) | 龙州 Longzhou (LLZ) | 4 | 22˚34′ N/106˚85′ E |
| 郁江 Yujiang (YJ) | 隆安 Long’an (LA) | 3 | 23˚17′ N/107˚69′ E |
| 黔江 Qianjiang (QJ) | 桂平 Guiping (GP) | 7 | 23˚39′ N/110˚07′ E |
| 韩江 Hanjiang (HJ) | 梅州 Meizhou (MZ) | 16 | 24˚29′ N/116˚11′ E |
| 韩江 Hanjiang (HJ) | 蕉岭 Jiaoling (JL) | 4 | 24˚65′ N/116˚17′ E |
| 韩江 Hanjiang (HJ) | 上杭 Shanghang (SH) | 4 | 25˚04′ N/116˚42′ E |
| 总计 Total | 111 |
Table 1 Sampling information of Hemibagrus guttatus samples
| 河流 River | 采集地 Sampling sites | 样本量 Sample size | 经纬度 Locality |
|---|---|---|---|
| 南盘江 Nanpanjiang (NPJ) | 西林 Xiling (XL) | 7 | 24˚39′ N/104˚28′ E |
| 柳江 Liujiang (LJ) | 宜州 Yizhou (YZ) | 8 | 24˚50′ N/108˚60′ E |
| 柳江 Liujiang (LJ) | 柳州 Liuzhou (LZZ) | 2 | 24˚31′ N/109˚42′ E |
| 柳江 Liujiang (LJ) | 柳城 Liucheng (LC) | 8 | 24˚65′ N/109˚26′ E |
| 桂江 Guijiang (GJ) | 昭平 Zhaoping (ZP) | 2 | 24˚17′ N/110˚80′ E |
| 桂江 Guijiang (GJ) | 平乐 Pingle (PL) | 4 | 24˚62′ N/110˚62′ E |
| 东江 Dongjiang (DJ) | 古竹 Guzhu (GZ) | 4 | 23˚51′ N/114˚70′ E |
| 东江 Dongjiang (DJ) | 惠州 Huizhou (HZ) | 2 | 23˚11′ N/114˚41′ E |
| 贺江 Hejiang (HEJ) | 南丰 Nanfeng (NF) | 5 | 23˚44′ N/111˚47′ E |
| 北江 Beijiang (BJ) | 韶关 Shaoguan (SG) | 4 | 24˚81′ N/113˚59′ E |
| 北江 Beijiang (BJ) | 连州 Lianzhou (LZ) | 8 | 24˚78′ N/112˚37′ E |
| 北江 Beijiang (BJ) | 西牛 Xiniu (XN) | 14 | 24˚17′ N/113˚11′ E |
| 红水河 Hongshuihe (HSH) | 都安 Du’an (DA) | 5 | 23˚93′ N/108˚10′ E |
| 郁江 Yujiang (YJ) | 龙州 Longzhou (LLZ) | 4 | 22˚34′ N/106˚85′ E |
| 郁江 Yujiang (YJ) | 隆安 Long’an (LA) | 3 | 23˚17′ N/107˚69′ E |
| 黔江 Qianjiang (QJ) | 桂平 Guiping (GP) | 7 | 23˚39′ N/110˚07′ E |
| 韩江 Hanjiang (HJ) | 梅州 Meizhou (MZ) | 16 | 24˚29′ N/116˚11′ E |
| 韩江 Hanjiang (HJ) | 蕉岭 Jiaoling (JL) | 4 | 24˚65′ N/116˚17′ E |
| 韩江 Hanjiang (HJ) | 上杭 Shanghang (SH) | 4 | 25˚04′ N/116˚42′ E |
| 总计 Total | 111 |
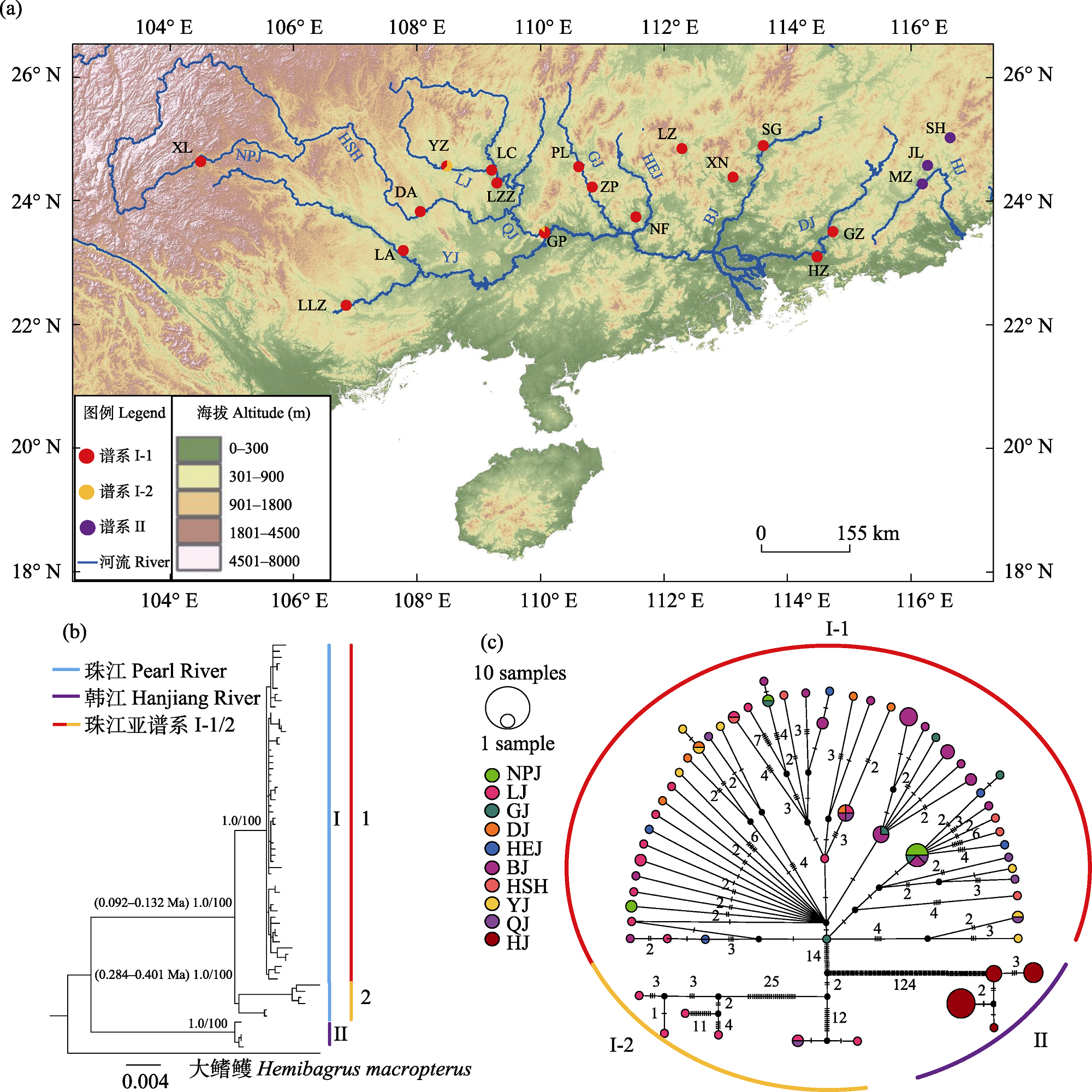
Fig. 1 Map of sample sites of Hemibagrus guttatus populations (a), phylogenetic tree (b) and haplotype network (c). In Figure a, the solid cycles indicate sample sites; the black and red fonts represent the abbreviations of sample sites and the river sections, respectively. In Figure b, the values in brackets indicate the divergence time, the values at left and right of slash represent the Bayesian posterior probability and support values of Maximum likelihood tree, respectively. In Figure c, the size of circle represents haplotype frequency and the numbers represent mutation steps. Sublineages I-1 and I-2 demarcate distinct evolutionary subdivisions, population codes refer to Table 1.
| 采样点 Sampling site | 样本大小 Sample size | 单倍型数 Number of haplotype | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 中性检测 Neutrality test | |
|---|---|---|---|---|---|---|
| Tajima’s D | Fu’s Fs | |||||
| 南盘江 NPJ | 7 | 3 | 0.667 (± 0.160) | 0.0004 (± 0.0001) | -0.367 | 3.708 |
| 柳江 LJ | 18 | 17 | 0.993 (± 0.021) | 0.0018 (± 0.0003) | -0.607 | -2.697 |
| 桂江 GJ | 6 | 6 | 1.000 (± 0.096) | 0.0005 (± 0.0001) | -0.644 | -1.522 |
| 东江 DJ | 6 | 6 | 1.000 (± 0.096) | 0.0005 (± 0.0001) | -0.315 | -1.522 |
| 贺江 HEJ | 5 | 5 | 1.000 (± 0.126) | 0.0006 (± 0.0001) | -0.770 | -0.542 |
| 北江 BJ | 27 | 15 | 0.942 (± 0.027) | 0.0004 (± 0.0001) | -1.707* | -3.177 |
| 红水河 HSH | 5 | 5 | 1.000 (± 0.126) | 0.0007 (± 0.0001) | -0.393 | -0.338 |
| 郁江 YJ | 7 | 7 | 1.000 (± 0.076) | 0.0007 (± 0.0001) | -0.957 | -1.528 |
| 黔江 QJ | 7 | 7 | 1.000 (± 0.076) | 0.0011 (± 0.0003) | -1.487* | -0.754 |
| 珠江 ZJ | 87 | 59 | 1.000 (± 0.003) | 0.0011 (± 0.0002) | -0.805 | -0.930 |
| 韩江 HJ | 24 | 4 | 0.641 (± 0.076) | 0.0002 (± 0.0002) | 1.526 | 3.312 |
| 总计 Total | 111 | 63 | 1.000 (± 0.003) | 0.002 (± 0.0005) | -0.986 | 2.243 |
Table 2 Detection of number of haplotype, haplotype diversity, nucleotide diversity and neutrality test of Hemibagrus guttatus. Population codes see Table 1.
| 采样点 Sampling site | 样本大小 Sample size | 单倍型数 Number of haplotype | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 中性检测 Neutrality test | |
|---|---|---|---|---|---|---|
| Tajima’s D | Fu’s Fs | |||||
| 南盘江 NPJ | 7 | 3 | 0.667 (± 0.160) | 0.0004 (± 0.0001) | -0.367 | 3.708 |
| 柳江 LJ | 18 | 17 | 0.993 (± 0.021) | 0.0018 (± 0.0003) | -0.607 | -2.697 |
| 桂江 GJ | 6 | 6 | 1.000 (± 0.096) | 0.0005 (± 0.0001) | -0.644 | -1.522 |
| 东江 DJ | 6 | 6 | 1.000 (± 0.096) | 0.0005 (± 0.0001) | -0.315 | -1.522 |
| 贺江 HEJ | 5 | 5 | 1.000 (± 0.126) | 0.0006 (± 0.0001) | -0.770 | -0.542 |
| 北江 BJ | 27 | 15 | 0.942 (± 0.027) | 0.0004 (± 0.0001) | -1.707* | -3.177 |
| 红水河 HSH | 5 | 5 | 1.000 (± 0.126) | 0.0007 (± 0.0001) | -0.393 | -0.338 |
| 郁江 YJ | 7 | 7 | 1.000 (± 0.076) | 0.0007 (± 0.0001) | -0.957 | -1.528 |
| 黔江 QJ | 7 | 7 | 1.000 (± 0.076) | 0.0011 (± 0.0003) | -1.487* | -0.754 |
| 珠江 ZJ | 87 | 59 | 1.000 (± 0.003) | 0.0011 (± 0.0002) | -0.805 | -0.930 |
| 韩江 HJ | 24 | 4 | 0.641 (± 0.076) | 0.0002 (± 0.0002) | 1.526 | 3.312 |
| 总计 Total | 111 | 63 | 1.000 (± 0.003) | 0.002 (± 0.0005) | -0.986 | 2.243 |
| 韩江 HJ | 南盘江 NPJ | 柳江 LJ | 桂江 GJ | 东江 DJ | 贺江 HEJ | 北江 BJ | 红水河 HSH | 郁江 YJ | 黔江 QJ | |
|---|---|---|---|---|---|---|---|---|---|---|
| 韩江 HJ | 1.01% | 1.03% | 1.01% | 1.01% | 1.02% | 1.13% | 1.02% | 1.02% | 1.01% | |
| 南盘江 NPJ | 0.978 | 0.13% | 0.04% | 0.05% | 0.04% | 0.20% | 0.05% | 0.06% | 0.07% | |
| 柳江 LJ | 0.913 | 0.111 | 0.14% | 0.13% | 0.14% | 0.29% | 0.15% | 0.14% | 0.15% | |
| 桂江 GJ | 0.976 | 0.000 | 0.072 | 0.05% | 0.05% | 0.20% | 0.06% | 0.06% | 0.08% | |
| 东江 DJ | 0.976 | 0.206 | 0.070 | 0.098 | 0.06% | 0.21% | 0.06% | 0.06% | 0.08% | |
| 贺江 HEJ | 0.976 | 0.000 | 0.055 | 0.000 | 0.086 | 0.21% | 0.06% | 0.07% | 0.08% | |
| 北江 BJ | 0.970 | 0.150 | 0.185 | 0.010 | 0.116 | 0.071 | 0.22% | 0.22% | 0.24% | |
| 红水河 HSH | 0.974 | 0.026 | 0.077 | 0.000 | 0.130 | 0.000 | 0.172 | 0.07% | 0.09% | |
| 郁江 YJ | 0.974 | 0.126 | 0.083 | 0.047 | 0.026 | 0.041 | 0.120 | 0.091 | 0.09% | |
| 黔江 QJ | 0.962 | 0.017 | 0.017 | 0.000 | 0.028 | 0.000 | 0.137 | 0.000 | 0.000 |
Table 3 Pairwise population F-statistics values (FST, below diagonal) and genetic distance (above diagonal) of Hemibagrus guttatus
| 韩江 HJ | 南盘江 NPJ | 柳江 LJ | 桂江 GJ | 东江 DJ | 贺江 HEJ | 北江 BJ | 红水河 HSH | 郁江 YJ | 黔江 QJ | |
|---|---|---|---|---|---|---|---|---|---|---|
| 韩江 HJ | 1.01% | 1.03% | 1.01% | 1.01% | 1.02% | 1.13% | 1.02% | 1.02% | 1.01% | |
| 南盘江 NPJ | 0.978 | 0.13% | 0.04% | 0.05% | 0.04% | 0.20% | 0.05% | 0.06% | 0.07% | |
| 柳江 LJ | 0.913 | 0.111 | 0.14% | 0.13% | 0.14% | 0.29% | 0.15% | 0.14% | 0.15% | |
| 桂江 GJ | 0.976 | 0.000 | 0.072 | 0.05% | 0.05% | 0.20% | 0.06% | 0.06% | 0.08% | |
| 东江 DJ | 0.976 | 0.206 | 0.070 | 0.098 | 0.06% | 0.21% | 0.06% | 0.06% | 0.08% | |
| 贺江 HEJ | 0.976 | 0.000 | 0.055 | 0.000 | 0.086 | 0.21% | 0.06% | 0.07% | 0.08% | |
| 北江 BJ | 0.970 | 0.150 | 0.185 | 0.010 | 0.116 | 0.071 | 0.22% | 0.22% | 0.24% | |
| 红水河 HSH | 0.974 | 0.026 | 0.077 | 0.000 | 0.130 | 0.000 | 0.172 | 0.07% | 0.09% | |
| 郁江 YJ | 0.974 | 0.126 | 0.083 | 0.047 | 0.026 | 0.041 | 0.120 | 0.091 | 0.09% | |
| 黔江 QJ | 0.962 | 0.017 | 0.017 | 0.000 | 0.028 | 0.000 | 0.137 | 0.000 | 0.000 |
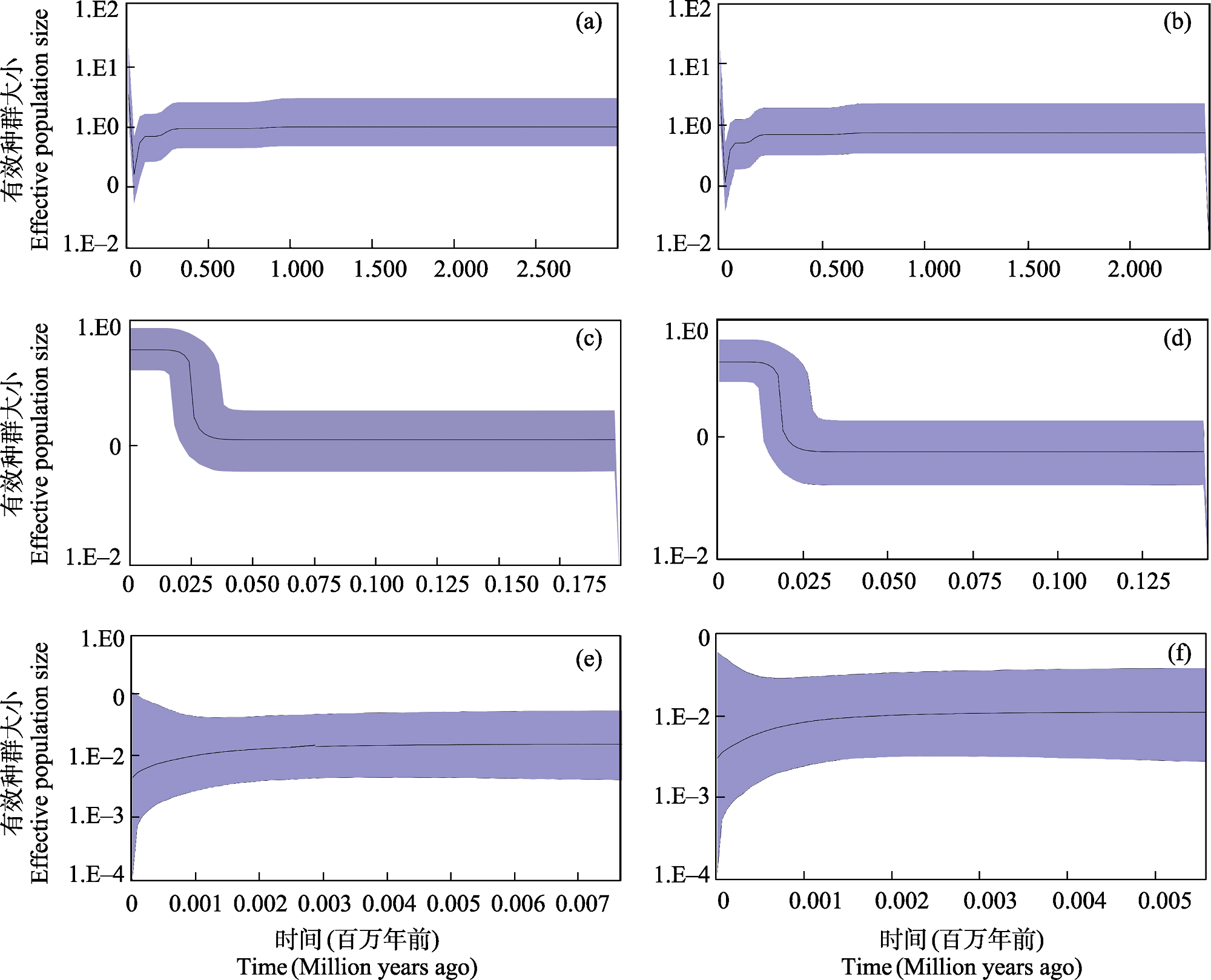
Fig. 2 Bayesian skyline plots of the overall population (a), the Pearl River population (c) and the Hanjiang River population (e) basedon the evolutionary rate of 0.62%, the overall population (b), the Pearl River population (d) and the Hanjiang River population (f) based on the evolutionary rate of 0.87%
| [1] | Billington N, Hebert PDN (1991) Mitochondrial DNA diversity in fishes and its implications for introductions. Canadian Journal of Fisheries and Aquatic Sciences, 48, 80-94. |
| [2] | Chen CW, Lin HW (1994) Geomorphology and climate evolution of the Hanjiang River delta. Journal of Hanshan Teachers College, (1), 7-14. (in Chinese) |
| [陈传五, 林海卫 (1994) 韩江三角洲地貌与气候的演变. 韩山师专学报, (1), 7-14.] | |
| [3] | Chen F, Lei H, Zheng HT, Wang WJ, Fang YH, Yang Z, Huang DM (2018) Impacts of cascade reservoirs on fishes in the mainstream of Pearl River and mitigation measures. Journal of Lake Sciences, 30, 1097-1108. (in Chinese with English abstract) |
| [陈锋, 雷欢, 郑海涛, 王文君, 方艳红, 杨钟, 黄道明 (2018) 珠江干流梯级开发对鱼类的影响与减缓对策. 湖泊科学, 30, 1097-1108.] | |
| [4] | Chen S, Jiao ZY, Zhu XP, Chen KC, Zheng GM, Chen YL (2006) Electrophoretic study of lactate dehydrogenase and esterase isozyme in Hemibagrus guttatus from the Xijiang River. Journal of Zhanjiang Ocean University, 6(26), 58-62. (in Chinese) |
| [陈赛, 焦宗垚, 朱新平, 陈昆慈, 郑光明, 陈永乐 (2006) 西江野生斑鳠乳酸脱氢酶和酯酶同工酶的电泳研究. 湛江海洋大学学报(自然科学), 6(26), 58-62.] | |
| [5] | Chen WT, Xiang DG, Gao S, Zhu SL, Wu Z, Li YF, Li J (2024) Whole-genome resequencing confirms the genetic effects of dams on an endangered fish Hemibagrus guttatus (Siluriformes: Bagridae): A case study in a tributary of the Pearl River. Gene, 895, 148000. |
| [6] |
Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM (2009) The last glacial maximum. Science, 325, 710-714.
DOI PMID |
| [7] |
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214.
PMID |
| [8] |
Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792-1797.
DOI PMID |
| [9] |
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [10] |
Fu YX (1996) Estimating the age of the common ancestor of a DNA sample using the number of segregating sites. Genetics, 144, 829-838.
DOI PMID |
| [11] | Gao F, Tan HY, Zhang M, Wang ZG, Wang KC, Chen XL (2020) Morphological variation analysis of Hemibagrus guttatus on main stream of Xijiang River in Guangxi. Southwest China Journal of Agricultural Sciences, 33, 1093-1100. (in Chinese with English abstract) |
| [高峰, 谭虹雨, 张曼, 王振光, 汪开成, 陈秀荔 (2020) 广西西江干流斑点半鲿形态差异分析. 西南农业学报, 33, 1093-1100.] | |
| [12] | Gao S, Li YF, Li J, Chen WT (2023) Genetic structure and demographic history of Mastacembelus armatus in southern China. South China Fisheries Science, 19(2), 42-49. (in Chinese with English abstract) |
| [高尚, 李跃飞, 李捷, 陈蔚涛 (2023) 华南地区大刺鳅的遗传结构与种群动态历史研究. 南方水产科学, 19(2), 42-49.] | |
| [13] |
Gascoyne M, Benjamin GJ, Schwarcz HP, Ford DC (1979) Sea level lowering during the Illinoian glaciation: Evidence from a Bahama “blue hole”. Science, 205, 806-808.
PMID |
| [14] | Gu S, Yi MR, He XB, Lin PS, Liu WH, Luo ZS, Lin HD, Yan YR (2021) Genetic diversity and population structure of cutlassfish (Lepturacanthus savala) along the coast of mainland China’s, as inferred by mitochondrial and microsatellite DNA markers. Regional Studies in Marine Science, 43, 101702. |
| [15] | Jiao ZY, Chen S, Zhu XP, Du HJ, Zheng GM, Chen KC (2007) Analysis of genetic diversity in Mystus guttatus from Xijiang branch of Pearl River. Journal of Agricultural Biotechnology, 15(1), 58-62. (in Chinese with English abstract) |
| [焦宗垚, 陈赛, 朱新平, 杜合军, 郑光明, 陈昆慈 (2007) 野生斑鳠西江地理群的遗传多样性分析. 农业生物技术学报, 15(1), 58-62.] | |
| [16] |
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111-120.
DOI PMID |
| [17] | Kuang TX, Shuai FM, Li XH, Chen WT, Lek S (2021) Genetic diversity and population structure of Hemibagrus guttatus (Bagridae, Siluriformes) in the larger subtropical Pearl River based on COI and Cyt b genes analysis. Annales De Limnologie, 57, 7. |
| [18] | Lan ZJ, Lin LF, Zhao J (2017) Genetic structure of Hemibarbus labeo and Hemibarbus medius in South China based on mtDNA COI and ND5 genes. Chinese Journal of Applied Ecology, 28, 1377-1386. (in Chinese with English abstract) |
|
[蓝昭军, 林龙峰, 赵俊 (2017) 基于COI和ND5序列的中国南部唇䱻和间䱻群体遗传结构. 应用生态学报, 28, 1377-1386.]
DOI |
|
| [19] |
Lanfear R, Calcott B, Ho SYW, Guindon S (2012) Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29, 1695-1701.
DOI PMID |
| [20] | Leigh JW, Bryant D (2015) Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110-1116. |
| [21] | Li HJ, Chu ZJ (2006) The biological character and breeding technology of Mystus guttatus. Journal of Anhui Agricultural Sciences, 34, 5882-5883, 5911. (in Chinese with English abstract) |
| [李红敬, 褚张杰 (2006) 斑鳠的生物学特性及养殖技术. 安徽农业科学, 34, 5882-5883, 5911.] | |
| [22] | Liang XX, Qing N, Yang KL, Wan CX, Zhao J, Chen XL (2010) Population genetic variation and phylogeography of Zacco platypus in Guangdong Province. Acta Hydrobiologica Sinica, 34, 806-814. (in Chinese with English abstract) |
| [梁晓旭, 庆宁, 杨柯林, 万彩霞, 赵俊, 陈湘粦 (2010) 广东地区宽鳍鱲种群遗传变异和亲缘地理. 水生生物学报, 34, 806-814.] | |
| [23] |
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [24] | Lin XZ, Hu YL, Wang RX, Li DM, Lin HS, Zha GC, Wen RS, Wu XQ (2023) Phytoplankton community structure and water quality assessment of the Chaozhou section of Hanjiang River. Journal of Hydroecology, 44(4), 52-60. (in Chinese with English abstract) |
| [林小植, 胡苑玲, 王瑞旋, 李冬梅, 林鸿生, 查广才, 温茹淑, 吴晓琼 (2023) 广东韩江潮州段浮游植物群落结构特征与水质评价. 水生态学杂志, 44(4), 52-60.] | |
| [25] | Liu W, Dai YG, Yuan ZX, Sun JJ, Liu L (2016) Genetic diversity of 2 economically important fishes in Siluriformes from Duliu River based on mtDNA D-loop sequences. Fisheries Science, 35, 386-392. (in Chinese with English abstract) |
| [刘伟, 代应贵, 袁振兴, 孙际佳, 刘丽 (2016) 基于线粒体DNA D-loop的都柳江鲇形目两种经济鱼类种群遗传多样性研究. 水产科学, 35, 386-392.] | |
| [26] | Meng GL, Li YY, Yang CT, Liu SL (2019) MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Research, 47, e63. |
| [27] | Pan TH (1985) Annals of Freshwater Fishes of Guangdong. Science and Technology Press, Guangdong. (in Chinese) |
| [潘炯华 (1985) 广东淡水鱼类志. 科技出版社, 广东.] | |
| [28] | Patel RK, Jain M (2012) NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE, 7, e30619. |
| [29] | Peng M, Han YQ, Wang DP, Shi J, Wu WJ, Li YS, Lei JJ, He AY (2020) Genetic diversity analysis of Ptychidio jordani in Xijiang River flowing through Guangxi Province based on mitochondrial Cytb gene sequence. South China Fisheries Science, 16(5), 10-18. (in Chinese with English abstract) |
| [彭敏, 韩耀全, 王大鹏, 施军, 吴伟军, 李育森, 雷建军, 何安尤 (2020) 基于线粒体Cytb基因序列的西江流域广西境内卷口鱼遗传多样性分析. 南方水产科学, 16(5), 10-18.] | |
| [30] | Qing N, Ma TF, Liang XX, Lin HD, Lu WH, Yan SL (2011) Population genetics and biogeography of Bufo melanostictus (Anura, Bufonidae) in South China. Acta Zootaxonomica Sinica, 36, 356-367. (in Chinese with English abstract) |
| [庆宁, 马天峰, 梁晓旭, 林弘都, 卢文华, 盐司橹 (2011) 华南地区黑眶蟾蜍的遗传变异和地理分化. 动物分类学报, 36, 356-367.] | |
| [31] | Rambaut A (2012) FigTree v1.4: Molecular Evolution, Phylogenetics and Epidemiology. University of Edinburgh, Institute of Evolutionary Biology, Edinburgh. |
| [32] | Ran GX, Dai YG, Yue XT (2012) Structure and genetic diversity of mtDNA D loop in the population of Onychostoma rara from the Xijiang River. Journal of Shanghai Ocean University, 21, 176-182. (in Chinese with English abstract) |
| [冉光鑫, 代应贵, 岳晓烔 (2012) 稀有白甲鱼西江种群mtDNA D环区的结构及遗传多样性. 上海海洋大学学报, 21, 176-182.] | |
| [33] |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574.
DOI PMID |
| [34] |
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312-1313.
DOI PMID |
| [35] |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
DOI PMID |
| [36] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725-2729. |
| [37] | Wang C, Zou Q, Yao DL, Zou JX, Zhou AG, Chen JT (2013) Preliminary study on artificial culture technology of wild Mystus guttatus in pond. Science Fish Farming, (9), 40-41. (in Chinese) |
| [王超, 邹青, 姚东林, 邹记兴, 周爱国, 陈金涛 (2013) 野生斑鳠池塘人工养殖技术初探. 科学养鱼, (9), 40-41.] | |
| [38] | Wu XM, Zhou LB, Peng FY (2010) Dressed rate and nutrition components in the muscle of spotted longbarbel catfish Mystus guttatus. Fisheries Science, 29, 425-428. (in Chinese with English abstract) |
| [吴小明, 周立斌, 彭飞影 (2010) 斑鳠的含肉率及肌肉营养成分分析. 水产科学, 29, 425-428.] | |
| [39] | Yang JP, Li C, Chen WT, Li YF, Li XH (2018) Genetic diversity and population demographic history of Ochetobius elongatus in the middle and lower reaches of the Xijiang River. Biodiversity Science, 26, 1289-1295. (in Chinese with English abstract) |
|
[杨计平, 李策, 陈蔚涛, 李跃飞, 李新辉 (2018) 西江中下游鳤的遗传多样性与种群动态历史. 生物多样性, 26, 1289-1295.]
DOI |
|
| [40] | Yang JQ, Hsu KC, Liu ZZ, Su LW, Kuo PH, Tang WQ, Zhou ZC, Liu D, Bao BL, Lin HD (2016) The population history of Garra orientalis (Teleostei: Cyprinidae) using mitochondrial DNA and microsatellite data with approximate Bayesian computation. BMC Evolutionary Biology, 16, 73. |
| [41] |
Yang L, He SP (2008) Phylogeography of the freshwater catfish Hemibagrus guttatus (Siluriformes, Bagridae): Implications for South China biogeography and influence of sea-level changes. Molecular Phylogenetics and Evolution, 49, 393-398.
DOI PMID |
| [42] | Zheng BS (1981) Annals of Freshwater Fishes of Guangxi. People’s Publishing House, Nanning. (in Chinese) |
| [郑葆珊 (1981) 广西淡水鱼类志. 广西人民出版社, 南宁.] |
| [1] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [2] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [3] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [4] | Congcong Lu, Qian Liu, Xiaolei Huang. Mitochondrial genome data of three aphid species [J]. Biodiv Sci, 2022, 30(7): 22204-. |
| [5] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [6] | Junjie Zhai, Huifeng Zhao, Guangshen Shang, Zhenjun Sun, Yufeng Zhang, Xing Wang. Advances in earthworm genomics: Based on whole genome and mitochondrial genome [J]. Biodiv Sci, 2022, 30(12): 22257-. |
| [7] | Yahong Zhang, Huixia Jia, Zhibin Wang, Pei Sun, Demei Cao, Jianjun Hu. Genetic diversity and population structure of Populus yunnanensis [J]. Biodiv Sci, 2019, 27(4): 355-365. |
| [8] | Xingtong Wu, Lu Chen, Minqiu Wang, Yuan Zhang, Xueying Lin, Xinyu Li, Hong Zhou, Yafeng Wen. Population structure and genetic divergence in Firmiana danxiaensis [J]. Biodiv Sci, 2018, 26(11): 1168-1179. |
| [9] | Qingqing Liu, Zhijun Dong. Population genetic structure of Gonionemus vertens based on the mitochondrial COI sequence [J]. Biodiv Sci, 2018, 26(11): 1204-1211. |
| [10] | Rong Li, Hang Sun. Phylofloristics: a case study from Yunnan, China [J]. Biodiv Sci, 2017, 25(2): 195-203. |
| [11] | Jie Liu, Yahuang Luo, Dezhu Li, Lianming Gao. Evolution and maintenance mechanisms of plant diversity in the Qinghai-Tibet Plateau and adjacent regions: retrospect and prospect [J]. Biodiv Sci, 2017, 25(2): 163-174. |
| [12] | Siyuan Ren, Ting Wang, Yan Zhu, Yongzhong Ye, Zhiliang Yuan, Cong Li, Na Pan, Luxin Li. Phylogenetic structure of individuals with different DBH sizes in a deciduous broad-leaved forest community in the temperate-subtropical ecological transition zone, China [J]. Biodiv Sci, 2014, 22(5): 574-582. |
| [13] | Min Xiong, Shuang Tian, Zhirong Zhang, Dengmei Fan, Zhiyong Zhang. Population genetic structure and conservation units of Sinomanglietia glauca (Magnoliaceae) [J]. Biodiv Sci, 2014, 22(4): 476-484. |
| [14] | Aihong Yang, Jinju Zhang, Hua Tian, Xiaohong Yao, Hongwen Huang. Microsatellite genetic diversity and fine-scale spatial genetic structure within a natural stand of Liriodendron chinense (Magnoliaceae) in Lanmushan, Duyun City, Guizhou Province [J]. Biodiv Sci, 2014, 22(3): 375-384. |
| [15] | Gangbiao Xu,Yan Liang,Yan Jiang,Xiongsheng Liu,Shangli Hu,Yufei Xiao,Bobo Hao. Genetic diversity and population structure of Bretschneidera sinensis, an endangered species [J]. Biodiv Sci, 2013, 21(6): 723-731. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()