



Biodiv Sci ›› 2014, Vol. 22 ›› Issue (3): 375-384. DOI: 10.3724/SP.J.1003.2014.14013 cstr: 32101.14.SP.J.1003.2014.14013
• Original Papers • Previous Articles Next Articles
Aihong Yang1,2, Jinju Zhang3, Hua Tian1, Xiaohong Yao1,*( ), Hongwen Huang1
), Hongwen Huang1
Received:2014-01-14
Accepted:2014-04-25
Online:2014-05-20
Published:2014-06-04
Contact:
Yao Xiaohong
Aihong Yang, Jinju Zhang, Hua Tian, Xiaohong Yao, Hongwen Huang. Microsatellite genetic diversity and fine-scale spatial genetic structure within a natural stand of Liriodendron chinense (Magnoliaceae) in Lanmushan, Duyun City, Guizhou Province[J]. Biodiv Sci, 2014, 22(3): 375-384.
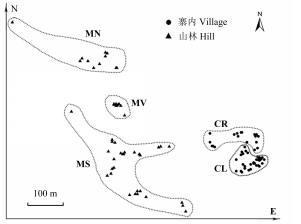
Fig. 1 Spatial distribution of Liriodendron chinense individu- als in Lanmushan population in Duyun, Guizhou. The five habitat fragments are circled with dash lines. CL, CR, MN, MS and MV refer to habitat fragments of left side of road in village, right side of road in village, north hill, south hill and hill valley, respectively. Different shapes represent two subpopulations, i.e. village subpopulation (●) and hill subpopulation (▲).
| 遗传参数 | N | A | AE | AR | Ho | HE | FIS |
|---|---|---|---|---|---|---|---|
| 年龄层 Age class | |||||||
| 幼树 Seedling | 19 | 3.462 | 2.492 | 3.462 | 0.526 | 0.582 | 0.097 |
| 小树 Sapling | 33 | 3.462 | 2.428 | 3.372 | 0.491 | 0.554 | 0.116 |
| 壮树 Adolescent | 29 | 3.615 | 2.469 | 3.520 | 0.502 | 0.564 | 0.111 |
| 成年树 Adult | 39 | 3.308 | 2.478 | 3.273 | 0.517 | 0.558 | 0.075 |
| 生境斑块 Habitat fragments | |||||||
| CL | 37 | 3.538 | 2.559 | 3.248 | 0.529 | 0.590 | 0.104 |
| CR | 26 | 3.385 | 2.462 | 3.255 | 0.497 | 0.549 | 0.097 |
| MN | 11 | 2.462 | 1.963 | 2.462 | 0.483 | 0.469 | -0.0031 |
| MS | 35 | 3.308 | 2.067 | 2.788 | 0.484 | 0.486 | 0.004 |
| MV | 11 | 2.846 | 2.302 | 2.846 | 0.559 | 0.564 | 0.009 |
| 物种水平 Species level | 120 | 4.000 | 2.508 | 4.000 | 0.508 | 0.563 | 0.099** |
Table 1 Summary of genetic diversity and fixation indices for four age-classes and five habitat fragments of Liriodendron chinense in Lanmushan population
| 遗传参数 | N | A | AE | AR | Ho | HE | FIS |
|---|---|---|---|---|---|---|---|
| 年龄层 Age class | |||||||
| 幼树 Seedling | 19 | 3.462 | 2.492 | 3.462 | 0.526 | 0.582 | 0.097 |
| 小树 Sapling | 33 | 3.462 | 2.428 | 3.372 | 0.491 | 0.554 | 0.116 |
| 壮树 Adolescent | 29 | 3.615 | 2.469 | 3.520 | 0.502 | 0.564 | 0.111 |
| 成年树 Adult | 39 | 3.308 | 2.478 | 3.273 | 0.517 | 0.558 | 0.075 |
| 生境斑块 Habitat fragments | |||||||
| CL | 37 | 3.538 | 2.559 | 3.248 | 0.529 | 0.590 | 0.104 |
| CR | 26 | 3.385 | 2.462 | 3.255 | 0.497 | 0.549 | 0.097 |
| MN | 11 | 2.462 | 1.963 | 2.462 | 0.483 | 0.469 | -0.0031 |
| MS | 35 | 3.308 | 2.067 | 2.788 | 0.484 | 0.486 | 0.004 |
| MV | 11 | 2.846 | 2.302 | 2.846 | 0.559 | 0.564 | 0.009 |
| 物种水平 Species level | 120 | 4.000 | 2.508 | 4.000 | 0.508 | 0.563 | 0.099** |
| CL | CR | MN | MS | MV | |
|---|---|---|---|---|---|
| CL | - | 0.007 | 0.050* | 0.028* | 0.048* |
| CR | 0.007 | - | 0.057* | 0.042* | 0.072* |
| MN | 0.096* | 0.102* | - | 0 | 0 |
| MS | 0.085* | 0.096* | 0 | - | 0 |
| MV | 0.056* | 0.069* | 0 | 0.011 | - |
Table 2 Pairwise genetic differentiation FST (below diagonal) and RST (above diagonal) for habitat fragments of Liriodendron chinense population in Lanmushan. Fragment codes see Fig. 1.
| CL | CR | MN | MS | MV | |
|---|---|---|---|---|---|
| CL | - | 0.007 | 0.050* | 0.028* | 0.048* |
| CR | 0.007 | - | 0.057* | 0.042* | 0.072* |
| MN | 0.096* | 0.102* | - | 0 | 0 |
| MS | 0.085* | 0.096* | 0 | - | 0 |
| MV | 0.056* | 0.069* | 0 | 0.011 | - |
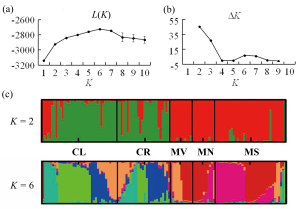
Fig. 2 Results of STRUCTURE analysis based on all the individuals in Lanmushan population using 13 polymorphic microsatellite loci. (a) Plotted the mean likelihood L(K); (b) ∆K value; and (c) Assignments proportion of each individual from all the five habitat fragments. Codes for habitat fragments correspond to those in Fig. 1.
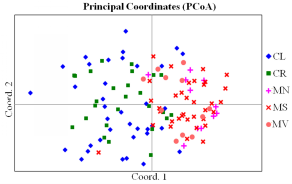
Fig. 3 Principal coordinate plot of genetic distance for the analyzed 120 individuals from five habitat fragments based on 13 SSR markers. The first two principal coordinates account for 13.58% and 9.43% of total genetic variation, respectively. Codes for habitat fragments correspond to those in Fig. 1.
| 组别 Group | N | P(r < permuted r) | F(1) | bF | Sp |
|---|---|---|---|---|---|
| 整体 Whole population | 120 | 0.001 | 0.0532 | -0.0085* | 0.0090 |
| 寨内 Village subpopulation | 63 | 0.001 | 0.0151 | -0.0066 | 0.0067 |
| 山林 Hill subpopulation | 57 | 0.013 | 0.0147 | -0.0052 | 0.0053 |
Table 3 Spatial genetic structure statistics for Liriodendron chinense population in Lanmushan
| 组别 Group | N | P(r < permuted r) | F(1) | bF | Sp |
|---|---|---|---|---|---|
| 整体 Whole population | 120 | 0.001 | 0.0532 | -0.0085* | 0.0090 |
| 寨内 Village subpopulation | 63 | 0.001 | 0.0151 | -0.0066 | 0.0067 |
| 山林 Hill subpopulation | 57 | 0.013 | 0.0147 | -0.0052 | 0.0053 |
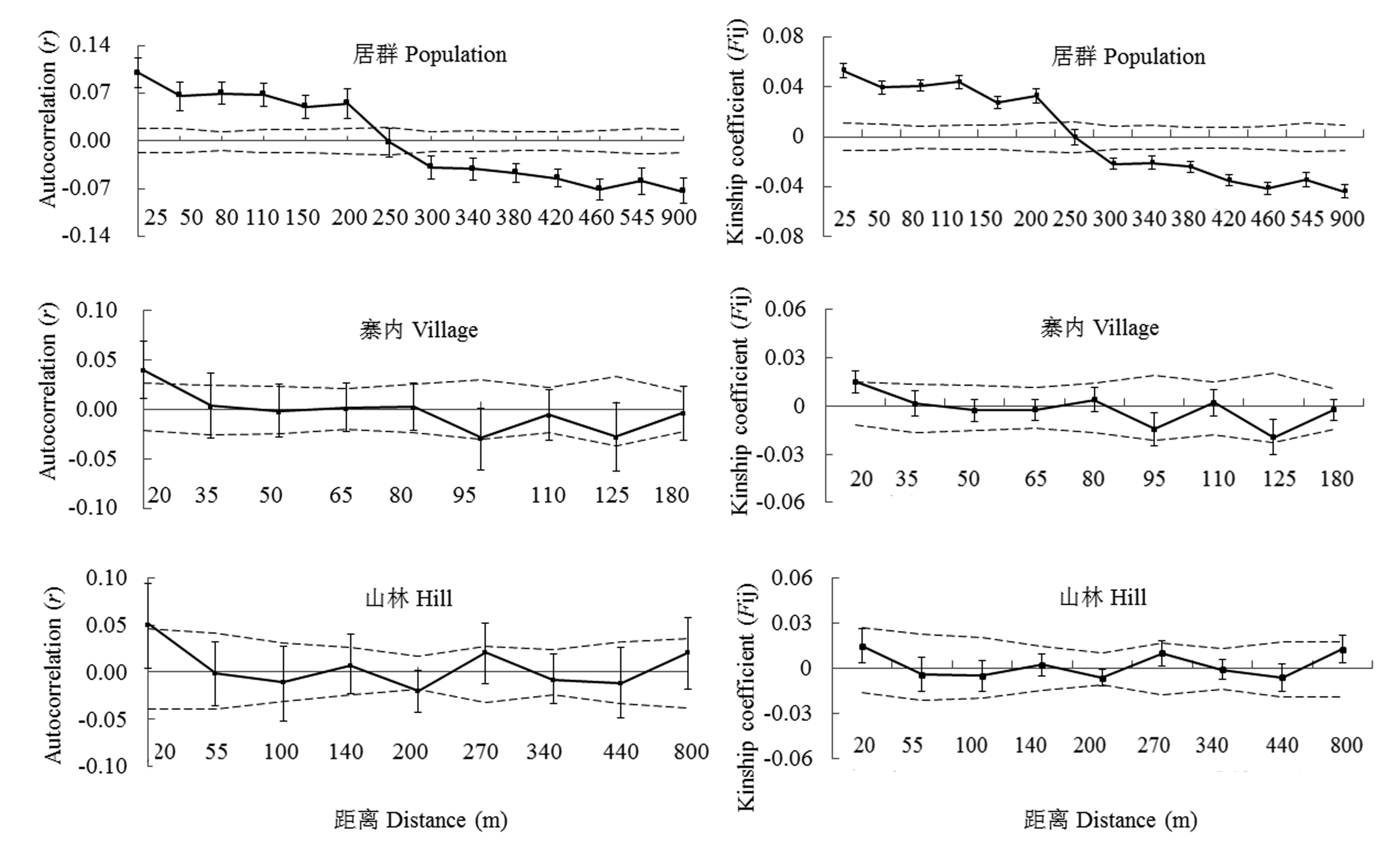
Fig. 4 Correlogram of coefficients for the population Lanmushan, village subpopulation and hill subpopulation. Autocorrelation coefficient (r) (left) and average kinship coefficients Fij (right) between pairs of individuals plotted as the classified geographical distance. Dashed lines represent 95% confidence intervals under the null hypothesis that genotypes are randomly distributed, and error bars delineate standard errors from Jackknife estimates.
| [1] | Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008) Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches.Molecular Ecology, 17, 5177-5188. |
| [2] | Bizoux JP, Dainou K, Bourland N, Hardy OJ, Heuertz M, Mahy G, Doucet JL (2009) Spatial genetic structure in Milicia excelsa (Moraceae) indicates extensive gene dispersal in a low-density wind-pollinated tropical tree.Molecular Ecology, 18, 4398-4408. |
| [3] | Born C, Hardy OJ, Chevallier MH, Ossari S, Attéké C, Wickings EJ, Hossaert-Mckey M (2008) Small-scale spatial genetic structure in the Central African rainforest tree species Aucoumea klaineana: a stepwise approach to infer the impact of limited gene dispersal, population history and habitat fragmentation.Molecular Ecology, 17, 2041-2050. |
| [4] | Busing RT (1995) Disturbance and the population dynamics of Liriodendron tulipifera.Journal of Ecology, 83, 45-53. |
| [5] | Cavers S, Degen B, Caron H, Lemes MR, Margis R, Salgueiro F, Lowe AJ (2005) Optimal sampling strategy for estimation of spatial genetic structure in tree populations.Heredity, 95, 281-289. |
| [6] | De-Lucas AI, Gonzalez-Martinez SC, Vendramin GG, Hidalgo E, Heuertz M (2009) Spatial genetic structure in continuous and fragmented populations of Pinus pinaster Aiton.Molecular Ecology, 18, 4564-4576. |
| [7] | Diniz-Filho JAF, De Campos Telles MP (2002) Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations.Conserva- tion Biology, 16, 924-935. |
| [8] | Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method.Conservation Genetics Resources, 4, 359-361. |
| [9] | Epperson BK (1992) Spatial structure of genetic variation within populations of forest trees.New Forests, 6, 257-278. |
| [10] | Epperson BK (1993) Recent advances in correlation studies of spatial patterns of genetic variation. In: Evolutionary Biology (eds Hecht M, MacIntyre R, Clegg M), pp. 95-155. Plenum Press, New York. |
| [11] | Epperson BK (1995) Spatial distributions of genotypes under isolation by distance.Genetics, 140, 1431-1440. |
| [12] | Escudero A, Iriondo JM, Torres ME (2003) Spatial analysis of genetic diversity as a tool for plant conservation.Biological Conservation, 113, 351-365. |
| [13] | Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3. 0): an integrated software package for population genetics data analysis.Evolutionary Bioinformatics from Online, 1, 47-50. |
| [14] | Fu LK (傅立国), Jin JM (金鉴明) (1992) The Red Data Book of China’s Plants: Rare and Endangered Species, Vol. 1 (中国植物红皮书: 稀有濒危植物·第一卷). Science Press, Beijing. (in Chinese) |
| [15] | Goudet J (2001) FSTAT, A Program to Estimate and Test Gene Diversities and Fixation Indices (version 2. 9. 3).. |
| [16] | Greene DF, Johnson EA (1989) A model of wind dispersal of winged or plumed seeds.Ecology, 70, 339-347. |
| [17] | Guillot G, Estoup A, Mortier F, Cosson JF (2005) A spatial statistical model for landscape genetics.Genetics, 170, 1261-1280. |
| [18] | Guo ZY (郭治友) (2003) Liriodendron chinense naturally distributes at Luosike forest for conservation of water nature sanctuary in Duyun City, Guizhou Province.Journal of Qiannan Normal College for Nationalities(黔南民族师范学院学报), 23(3), 34-36. (in Chinese with English abstract) |
| [19] | Guo ZY (郭治友), Xiao GX (肖国学), Zhao H (赵洪), Wu WD (吴卫东) (2008) Research on population ecology of Liriodendron chinense in Luosike Mountain in Duyun City.Journal of Anhui Agriculture Science(安徽农业科学), 36, 9970-9972. (in Chinese with English abstract) |
| [20] | Hao RM (郝日明), He SA (贺善安), Tang SJ (汤诗杰), Wu SP (伍寿彭) (1995) Geographical distribution of Liriodendron chinense in China and its significance.Journal of Plant Resources and Environment(植物资源与环境), 4, 1-6. (in Chinese with English abstract) |
| [21] | Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier MH, Doligez A, Dutech C, Kremer A, Latouche-Hallé C, Troispoux V, Veron V, Degen B (2006) Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species.Molecular Ecology, 15, 559-571. |
| [22] | Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels.Molecular Ecology Notes, 2, 618-620. |
| [23] | He R, Wang J, Huang H (2012) Long-distance gene dispersal inferred from spatial genetic structure in Handeliodendron bodinieri, an endangered tree from karst forest in southwest China.Biochemical Systematics and Ecology, 44, 295-302. |
| [24] | Heinken T, Weber E (2013) Consequences of habitat fragmentation for plant species: Do we know enough? Perspectives in Plant Ecology, Evolution and Systematics, 15, 205-216. |
| [25] | Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation.Conservation Biology, 21, 823-831. |
| [26] | Huang JQ (黄坚钦) (1998) Embryology reasons for lower seed-setting in Liriodendron chinense.Journal of Zhejiang Forestry College(浙江林学院学报), 15, 269-273. (in Chinese with English abstract) |
| [27] | Huang SQ (黄双全), Guo YH (郭友好) (2000) Pollination environment and sex allocation in Liriodendron chinense.Acta Ecologica Sinica(生态学报), 20, 49-52. (in Chinese with English abstract) |
| [28] | Jones FA, Hamrick JL, Peterson CJ, Squiers ER (2006) Inferring colonization history from analyses of spatial genetic structure within populations of Pinus strobus and Quercus rubra.Molecular Ecology, 15, 851-861. |
| [29] | Jump AS, Penuelas J (2006) Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proceedings of the National Academy of Sciences, USA, 103, 8096-8100. |
| [30] | Kalisz S, Nason JD, Hanzawa FM, Tonsor SJ (2001) Spatial population genetic structure in Trillium grandiflorum: the roles of dispersal, mating, history, and selection.Evolution, 55, 1560-1568. |
| [31] | Kramer AT, Ison JL, Ashley MV, Howe HF (2008) The paradox of forest fragmentation genetics.Conservation Biology, 22, 878-885. |
| [32] | Krauss SL, Koch JM (2004) Rapid genetic delineation of provenance for plant community restoration.Journal of Applied Ecology, 41, 1162-1173. |
| [33] | Leite FAB, Brandão RL, de Oliveira Buzatti RS, de Lemos-Filho JP, Lovato MB (2013) Fine-scale genetic structure of the threatened rosewood Dalbergia nigra from the Atlantic forest: comparing saplings versus adults and small fragment versus continuous forest.Tree Genetics and Genomes, 10, 307-316 |
| [34] | Levin DA, Kerster H (1969) Density-dependent gene dispersal in Liatris.The American Naturalist, 103, 61-74. |
| [35] | Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants.Annual Review of Ecology and Systematics, 27, 237-277. |
| [36] | Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae).American Journal of Botany, 82, 1420-1425. |
| [37] | Lowe AJ, Boshier D, Ward M, Bacles CF, Navarro C (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees.Heredity, 95, 255-273. |
| [38] | Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics.Trends in Ecology and Evolution, 18, 189-197. |
| [39] | Nathan R, Katul GG (2005) Foliage shedding in deciduous forests lifts up long-distance seed dispersal by wind.Proceedings of the National Academy of Sciences, USA, 102, 8251-8256. |
| [40] | Nathan R, Katul GG, Horn HS, Thomas SM, Oren R, Avissar R, Pacala SW, Levin SA (2002) Mechanisms of long- distance dispersal of seeds by wind.Nature, 418, 409-413. |
| [41] | Pandey M, Gailing O, Hattemer HH, Finkeldey R (2012) Fine-scale spatial genetic structure of sycamore maple (Acer pseudoplatanus L.).European Journal of Forest Research, 131, 739-746. |
| [42] | Pandey M, Geburek T (2011) Fine-scale genetic structure and gene flow in a semi-isolated population of a tropical tree, Shorea robusta Gaertn. (Dipterocarpaceae).Current Science, 101, 293-299. |
| [43] | Parks CR, Wendel JF (1990) Molecular divergence between Asian and North American species of Liriodendron (Magnoliaceae) with implications for interpretation of fossil floras.American Journal of Botany, 77, 1243-1256. |
| [44] | Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research.Molecular Ecology Notes, 6, 288-295. |
| [45] | Peakall ROD, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update.Bioinformatics, 28, 2537-2539. |
| [46] | Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data.Genetics, 155, 945-959. |
| [47] | Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure.Molecular Ecology Notes, 4, 137-138. |
| [48] | Sala OE, Chapin FS, Armesto JJ, Berlow E, Bloomfield J, Dirzo R, Huber-Sanwald E, Huenneke LF, Jackson RB, Kinzig A (2000) Global biodiversity scenarios for the year 2100.Science, 287, 1770-1774. |
| [49] | Slatkin M (1987) Gene flow and the geographic structure of natural populations.Science, 236, 787-792. |
| [50] | Smouse PE, Peakall ROD (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure.Heredity, 82, 561-573. |
| [51] | Sokal RR, Wartenberg DE (1983) A test of spatial autocorrelation analysis using an isolation-by-distance model.Genetics, 105, 219-237. |
| [52] | Su H, Qu LJ, He K, Zhang Z, Wang J, Chen Z, Gu H (2003) The Great Wall of China: a physical barrier to gene flow?Heredity, 90, 212-219. |
| [53] | Sun YG (孙亚光), Li HG (李火根) (2007) The paternity analysis for open-pollination progenies of Liriodendron L. using SSR markers.Chinese Bulletin of Botany(植物学通报), 24, 590-596. (in Chinese with English abstract) |
| [54] | Tang CQ, Yang Y, Ohsawa M, Momohara A, Mu J, Robertson K (2013) Survival of a Tertiary relict species, Liriodendron chinense (Magnoliaceae), in southern China, with special reference to village fengshui forests.American Journal of Botany, 100, 2112-2119. |
| [55] | Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations.Molecular Ecology, 13, 921-935. |
| [56] | Wang R, Compton SG, Chen XY (2011) Fragmentation can increase spatial genetic structure without decreasing pollen-mediated gene flow in a wind-pollinated tree.Molecular Ecology, 20, 4421-4432. |
| [57] | Wang R, Compton SG, Shi YS, Chen XY (2012) Fragmentation reduces regional-scale spatial genetic structure in a wind-pollinated tree because genetic barriers are removed.Ecology and Evolution, 2, 2250-2261. |
| [58] | White GM, Boshier DH, Powell W (2002) Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini.Proceedings of the National Academy of Sciences, USA, 99, 2038-2042. |
| [59] | Wright S (1943) Isolation by distance.Genetics, 28, 114. |
| [60] | Yang AH, Zhang JJ, Tian H, Yao XH (2012) Characterization of 39 novel EST-SSR markers for Liriodendron tulipifera and cross-species amplification in L. chinense (Magnolia- ceae).American Journal of Botany, 99, e460-e464. |
| [61] | Yao X, Ye Q, Kang M, Huang H (2007) Microsatellite analysis reveals interpopulation differentiation and gene flow in the endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China.New Phytologist, 176, 472-480. |
| [62] | Yao XH, Zhang JJ, Ye QG, Huang HW (2008) Characterization of 14 novel microsatellite loci in the endangered Liriodendron chinense (Magnoliaceae) and cross-species amplification in closely related taxa.Conservation Genetics, 9, 483-485. |
| [63] | Yao XH, Zhang JJ, Ye QG, Huang HW (2011) Fine-scale spatial genetic structure and gene flow in a small, fragmented population of Sinojackia rehderiana (Styracaceae), an endangered tree species endemic to China.Plant Biology, 13, 401-410. |
| [64] | Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants.Trends in Ecology and Evolution, 11, 413-418. |
| [65] | Zhou J (周坚), Fan RW (樊汝汶) (1999) Pollination of Liriodendron chinense.Chinese Bulletin of Botany(植物学通报), 16, 75-79. (in Chinese with English abstract) |
| [1] | Murong Yi, Ping Lu, Yong Peng, Yong Tang, Jiuheng Xu, Haoping Yin, Luyang Zhang, Xiaodong Weng, Mingxiao Di, Juan Lei, Chenqi Lu, Rujun Cao, Nianhua Dai, Deyang Zhan, Mei Tong, Zhiming Lou, Yonggang Ding, Jing Chai, Jing Che. Population status and habitat of Critically Endangered Jiangxi giant salamander (Andrias jiangxiensis) [J]. Biodiv Sci, 2025, 33(4): 24145-. |
| [2] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [5] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [6] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [7] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [8] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [9] | Kunming Zhao, Shengbin Chen, Xifu Yang. Investigation of the diversity of mammals and birds and the activity rhythm of dominant species using camera trapping in a fragmented forest in the Dujiangyan region, Sichuan Province [J]. Biodiv Sci, 2023, 31(6): 22529-. |
| [10] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [11] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [12] | Chang Cai, Xue Zhang, Chen Zhu, Yuhao Zhao, Gexia Qiao, Ping Ding. Nested assemblages of aphid species in the Thousand Island Lake: The importance of island area and host plant diversity [J]. Biodiv Sci, 2023, 31(12): 23183-. |
| [13] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [14] | Shuanggui Wang, Zhihong Guo, Bojian Gu, Tianti Li, Yubing Su, Bocheng Ma, Hongxin Guan, Qiaowen Huang, Fang Wang, Zhuojin Zhang. Habitat use of the North China leopard (Panthera pardus japonensis) in the Liupanshan Mountains and its implications for conservation planning [J]. Biodiv Sci, 2022, 30(9): 22342-. |
| [15] | Yunrui Ji, Xuelei Wei, Guofeng Zhang, Minggui Xiang, Yongchao Wang, Renhu Gong, Yang Hu, Diqiang Li, Fang Liu. Diversity and composition of bird species in the Hubei Wufeng Houhe National Nature Reserve [J]. Biodiv Sci, 2022, 30(7): 21475-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()