



Biodiv Sci ›› 2022, Vol. 30 ›› Issue (12): 22257. DOI: 10.17520/biods.2022257 cstr: 32101.14.biods.2022257
Special Issue: 土壤生物与土壤健康; 有机农业
• Reviews • Previous Articles Next Articles
Junjie Zhai1,2, Huifeng Zhao3, Guangshen Shang1,2, Zhenjun Sun1, Yufeng Zhang3,*( ), Xing Wang1,2,*(
), Xing Wang1,2,*( )
)
Received:2022-05-11
Accepted:2022-10-01
Online:2022-12-20
Published:2022-12-07
Contact:
*E-mail: qqzyf123@126.com; swwangxing@cau.edu.cn
Junjie Zhai, Huifeng Zhao, Guangshen Shang, Zhenjun Sun, Yufeng Zhang, Xing Wang. Advances in earthworm genomics: Based on whole genome and mitochondrial genome[J]. Biodiv Sci, 2022, 30(12): 22257.
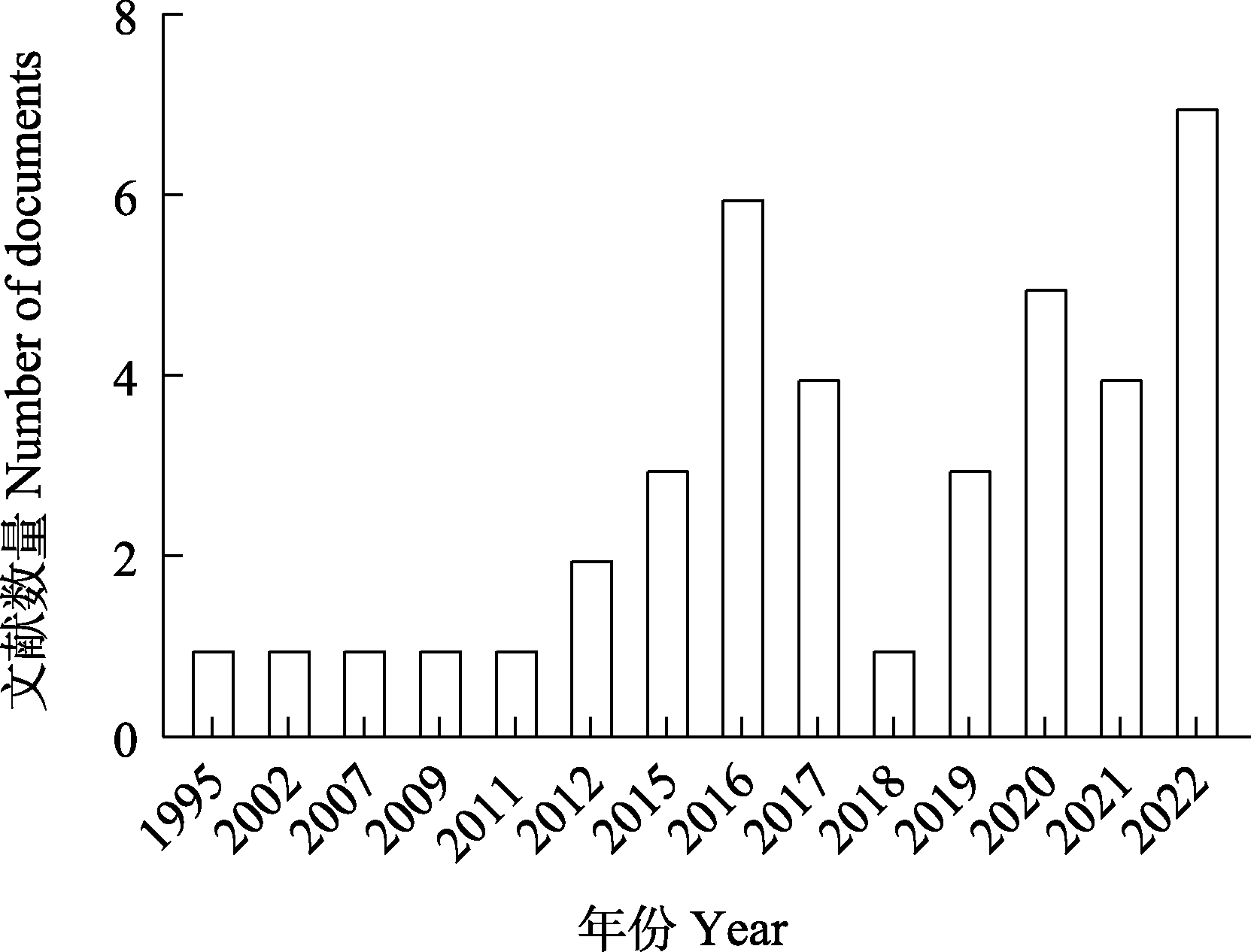
Fig. 1 Number of published papers on earthworm genomics. Literature source: Web of Science Core Collection; Publication years: 1995?2022; Search keywords: Earthworm, Genome.
| 蚯蚓 Earthworm | 基因组大小 Genome length | 染色体数量 Number of chromosomes | 染色体倍体 Chromosome ploidy | GC含量 GC content (%) | 蛋白编码基因数 Number of protein-coding gene | 测序组装技术 Sequencing assembly technology | 参考文献 References |
|---|---|---|---|---|---|---|---|
| 赤子爱胜蚓 Eisenia foetida | 1.05 Gb | 22 | 2 | 28.6 | ? | Illumina HiSeq 2000 2 × 100PE | 2015 |
| 赤子爱胜蚓 E. foetida | 1.47 Gb | 22 | 2 | 40 | ? | Illumina HiSeq 2500 | 2018 |
| 通俗腔蚓 Metaphire vulgaris | 728.6 Mb | 41 | 2 | 40 | ? | PromethION Single Molecule Platform, Illumina NovaSeq Sequencing Platform, Hi-C | 2020 |
| 安德爱胜蚓 Eisenia andrei | 1.3 Gb | 22 | 2 | 35?45 | 31,817 | PacBio三代测序 PacBio RS Platform, Hi-C | 2020 |
| 皮质远盲蚓 Amynthas corticis | 1.2 Gb | 42 | 3 | 40.34 | 29,256 | PacBio三代测序 PacBio SMRT Sequencing, Hi-C | 2021 |
Table 1 Basic information of whole genome sequencing in earthworm
| 蚯蚓 Earthworm | 基因组大小 Genome length | 染色体数量 Number of chromosomes | 染色体倍体 Chromosome ploidy | GC含量 GC content (%) | 蛋白编码基因数 Number of protein-coding gene | 测序组装技术 Sequencing assembly technology | 参考文献 References |
|---|---|---|---|---|---|---|---|
| 赤子爱胜蚓 Eisenia foetida | 1.05 Gb | 22 | 2 | 28.6 | ? | Illumina HiSeq 2000 2 × 100PE | 2015 |
| 赤子爱胜蚓 E. foetida | 1.47 Gb | 22 | 2 | 40 | ? | Illumina HiSeq 2500 | 2018 |
| 通俗腔蚓 Metaphire vulgaris | 728.6 Mb | 41 | 2 | 40 | ? | PromethION Single Molecule Platform, Illumina NovaSeq Sequencing Platform, Hi-C | 2020 |
| 安德爱胜蚓 Eisenia andrei | 1.3 Gb | 22 | 2 | 35?45 | 31,817 | PacBio三代测序 PacBio RS Platform, Hi-C | 2020 |
| 皮质远盲蚓 Amynthas corticis | 1.2 Gb | 42 | 3 | 40.34 | 29,256 | PacBio三代测序 PacBio SMRT Sequencing, Hi-C | 2021 |
| 分类单元 Taxa | GenBank登录号 GenBank accession no. | 参考文献 References |
|---|---|---|
| 正蚓科 Lumbricidae | ||
| Aporrectodea caliginosa | CM035405.1 | 未发表 Unpublished |
| A. rosea | NC_046733.1 | 2019 |
| A. tuberculata | OL840316?OL840317, OM687883?OM687886 | 2022 |
| A. trapezoides | OM687887 | 2022 |
| Bimastos parvus | MZ857199.1 | 2021 |
| Dendrobaena octaedra | MZ857197.1 | 2021 |
| Eisenia andrei | MZ857198.1 | 2021 |
| E. nordenskioldi | MZ857200.1 OL840314?OL840315 OM687887?OM687890 | 2021 2022 2022 |
| Lumbricus rubellus | MN102127.1 | 2019 |
| L. terrestris | NC_001673.1 | 1995 |
| Octolasion tyrtaeum | MZ857201.1 | 2021 |
| Pontoscolex corethrurus | NC_034783.1 | 未发表 Unpublished |
| 巨蚓科 Megascolecidae | ||
| Amynthas aspergillus | NC_025292.1 | 2016b |
| A. carnosus | KT429008.1 | 2016c |
| A. corticis | KM199290.1 | 2015 |
| A. cucullatus | KT429012.1 | 2016c |
| A. gracilis | KP688582.1 | 2015 |
| A. hupeiensis | KT429009.1 | 2016c |
| A. instabilis | KT429007.1 | 2016c |
| A. jiriensis | NC_029879.1 | 2017 |
| A. longisiphonus | KM199289.1 | 2015 |
| A. moniliatus | KT429020.1 | 2016c |
| A. morrisi | KT429011.1 | 2016c |
| A. pectiniferus | KT429018.1 | 2016c |
| A. redactus | KT429010.1 | 2016c |
| A. robustus | KT429019.1 | 2016c |
| A. rongshuiensis | KT429014.1 | 2016c |
| A. seungpanensis | OL321943.1 | 未发表 Unpublished |
| A. spatiosus | KT429013.1 | 2016c |
| A. triastriatus | KT429016.1 | 2016c |
| A. yunoshimensis | LC573969.1 | 2021 |
| Duplodicodrilus schmardae | KT429015.1 | 2016c |
| Metaphire californica | KP688581.1 | 2015 |
| M. guillelmi | KT429017.1 | 2016c |
| M. hilgendorfi | LC573968.1 | 2021 |
| M. vulgaris | NC_023836.1 | 2016a |
| Perionyx excavatus | NC_009631.1 | 未发表 Unpublished |
| Tonoscolex birmanicus | KF425518.1 | 2015 |
| 链胃蚓科 Moniligastridae | ||
| Drawida gisti | NC_058285.1 | 2020 |
| D. japonica | KM199288.1 | 2016c |
| D. ghilarovi | OL840312?OL840313 | 2022 |
Table 2 Taxonomic distribution of earthworm species based on mitochondriai genomes
| 分类单元 Taxa | GenBank登录号 GenBank accession no. | 参考文献 References |
|---|---|---|
| 正蚓科 Lumbricidae | ||
| Aporrectodea caliginosa | CM035405.1 | 未发表 Unpublished |
| A. rosea | NC_046733.1 | 2019 |
| A. tuberculata | OL840316?OL840317, OM687883?OM687886 | 2022 |
| A. trapezoides | OM687887 | 2022 |
| Bimastos parvus | MZ857199.1 | 2021 |
| Dendrobaena octaedra | MZ857197.1 | 2021 |
| Eisenia andrei | MZ857198.1 | 2021 |
| E. nordenskioldi | MZ857200.1 OL840314?OL840315 OM687887?OM687890 | 2021 2022 2022 |
| Lumbricus rubellus | MN102127.1 | 2019 |
| L. terrestris | NC_001673.1 | 1995 |
| Octolasion tyrtaeum | MZ857201.1 | 2021 |
| Pontoscolex corethrurus | NC_034783.1 | 未发表 Unpublished |
| 巨蚓科 Megascolecidae | ||
| Amynthas aspergillus | NC_025292.1 | 2016b |
| A. carnosus | KT429008.1 | 2016c |
| A. corticis | KM199290.1 | 2015 |
| A. cucullatus | KT429012.1 | 2016c |
| A. gracilis | KP688582.1 | 2015 |
| A. hupeiensis | KT429009.1 | 2016c |
| A. instabilis | KT429007.1 | 2016c |
| A. jiriensis | NC_029879.1 | 2017 |
| A. longisiphonus | KM199289.1 | 2015 |
| A. moniliatus | KT429020.1 | 2016c |
| A. morrisi | KT429011.1 | 2016c |
| A. pectiniferus | KT429018.1 | 2016c |
| A. redactus | KT429010.1 | 2016c |
| A. robustus | KT429019.1 | 2016c |
| A. rongshuiensis | KT429014.1 | 2016c |
| A. seungpanensis | OL321943.1 | 未发表 Unpublished |
| A. spatiosus | KT429013.1 | 2016c |
| A. triastriatus | KT429016.1 | 2016c |
| A. yunoshimensis | LC573969.1 | 2021 |
| Duplodicodrilus schmardae | KT429015.1 | 2016c |
| Metaphire californica | KP688581.1 | 2015 |
| M. guillelmi | KT429017.1 | 2016c |
| M. hilgendorfi | LC573968.1 | 2021 |
| M. vulgaris | NC_023836.1 | 2016a |
| Perionyx excavatus | NC_009631.1 | 未发表 Unpublished |
| Tonoscolex birmanicus | KF425518.1 | 2015 |
| 链胃蚓科 Moniligastridae | ||
| Drawida gisti | NC_058285.1 | 2020 |
| D. japonica | KM199288.1 | 2016c |
| D. ghilarovi | OL840312?OL840313 | 2022 |
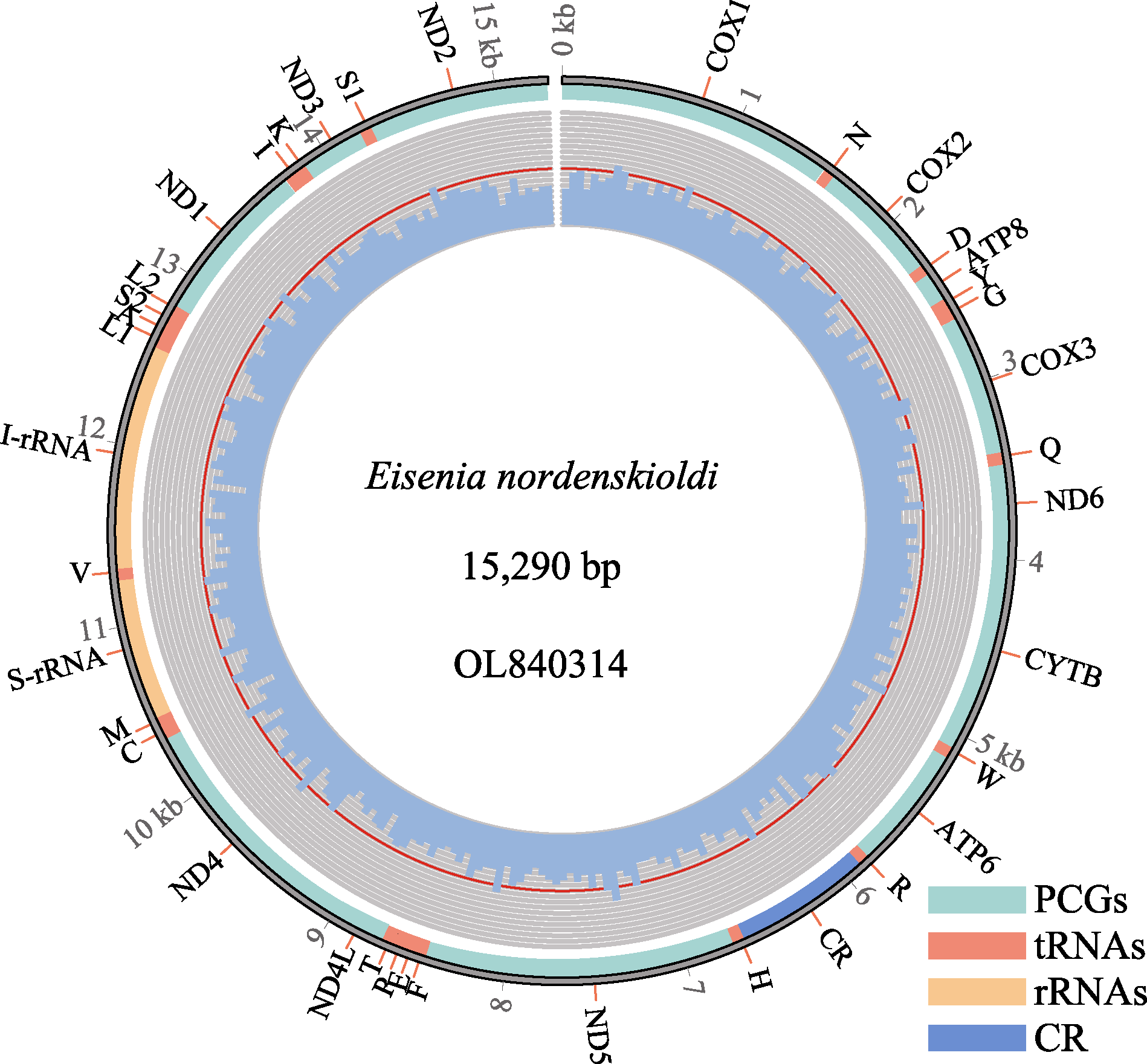
Fig. 3 The mitochondrial genome of Eisenia nordenskioldi. Cyan color represents protein-coding genes, red color represents transporter RNA, orange color represents ribosomal RNA, and blue color represents the control region.
| [1] |
Ahmed N, Al-Mutairi KA (2022) Earthworms effect on microbial population and soil fertility as well as their interaction with agriculture practices. Sustainability, 14, 7803.
DOI URL |
| [2] |
Anderson C, Cunha L, Sechi P, Kille P, Spurgeon D (2017) Genetic variation in populations of the earthworm, Lumbricus rubellus, across contaminated mine sites. BMC Genetics, 18, 97.
DOI PMID |
| [3] |
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA (2016) Harnessing the power of RADseq for ecological and evolutionary genomics. Nature Reviews Genetics, 17, 81-92.
DOI PMID |
| [4] | Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE, 3, e3376. |
| [5] |
Bertrand M, Barot S, Blouin M, Whalen J, Oliveira T, Roger-Estrade J (2015) Earthworm services for cropping systems. A review. Agronomy for Sustainable Development, 35, 553-567.
DOI URL |
| [6] | Bhambri A, Dhaunta N, Patel SS, Hardikar M, Bhatt A, Srikakulam N, Shridhar S, Vellarikkal S, Pandey R, Jayarajan R, Verma A, Kumar V, Gautam P, Khanna Y, Khan JA, Fromm B, Peterson KJ, Scaria V, Sivasubbu S, Pillai B (2018) Large scale changes in the transcriptome of Eisenia fetida during regeneration. PLoS ONE, 13, e0204234. |
| [7] |
Bhat SA, Singh J, Vig AP (2018) Earthworms as organic waste managers and biofertilizer producers. Waste and Biomass Valorization, 9, 1073-1086.
DOI URL |
| [8] |
Boore JL, Brown WM (1995) Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics, 141, 305-319.
DOI PMID |
| [9] | Bozorgi F, Seiedy M, Malek M, Aira M, Pérez-Losada M, Domínguez J (2019) Multigene phylogeny reveals a new Iranian earthworm genus (Lumbricidae: Philomontanus) with three new species. PLoS ONE, 14, e0208904. |
| [10] |
Conrado AC, Arruda H, Stanton DWG, James SW, Kille P, Brown G, Silva E, Dupont L, Taheri S, Morgan AJ, Simões N, Rodrigues A, Montiel R, Cunha L (2017) The complete mitochondrial DNA sequence of the pantropical earthworm Pontoscolex corethrurus (Rhinodrilidae, Clitellata): Mitogenome characterization and phylogenetic positioning. ZooKeys, 688, 1-13.
DOI URL |
| [11] | Csuzdi CS (2012) Earthworm species, a searchable database. Opuscula Zoologica Budapest, 43(41), 97-99. |
| [12] | Darwin C (1892) The Formation of Vegetable Mould, through the Action of Worms: With Observations on Their Habits. John Murray, London. |
| [13] | Ding SY, Lin XT, He SE (2019) Earthworms: A source of protein. Journal of Food Science and Engineering, 9, 159-170. |
| [14] |
Giska I, Sechi P, Babik W (2015) Deeply divergent sympatric mitochondrial lineages of the earthworm Lumbricus rubellus are not reproductively isolated. BMC Evolutionary Biology, 15, 217.
DOI URL |
| [15] |
Gong P, Perkins EJ (2016) Earthworm toxicogenomics: A renewed genome-wide quest for novel biomarkers and mechanistic insights. Applied Soil Ecology, 104, 12-24.
DOI URL |
| [16] | Hong Y, Kim MJ, Wang AR, Kim I (2017) Complete mitochondrial genome of the earthworm, Amynthas jiriensis (Clitellata: Megascolecidae). Mitochondrial DNA Part A, 28, 163-164. |
| [17] |
Jiang JB, Qiu JP (2018) Origin and evolution of earthworms belonging to the family Megascolecidae in China. Biodiversity Science, 26, 1074-1082. (in Chinese with English abstract)
DOI |
|
[ 蒋际宝, 邱江平 (2018) 中国巨蚓科蚯蚓的起源与演化. 生物多样性, 26, 1074-1082.]
DOI |
|
| [18] | Jin F, Zhou ZL, Guo Q, Liang ZW, Yang RY, Jiang JB, He YL, Zhao Q, Zhao Q (2020) High-quality genome assembly of Metaphire vulgaris. PeerJ, 8, e10313. |
| [19] |
Li Y, Tang H, Hu Y, Wang X, Ai X, Tang L, Matthew C, Cavanagh J, Qiu J (2016) Enrofloxacin at environmentally relevant concentrations enhances uptake and toxicity of cadmium in the earthworm Eisenia fetida in farm soils. Journal of Hazardous Materials, 308, 312-320.
DOI URL |
| [20] |
Li Y, Wang X, Sun Z (2020) Ecotoxicological effects of petroleum-contaminated soil on the earthworm Eisenia fetida. Journal of Hazardous Materials, 393, 122384.
DOI URL |
| [21] | Liu C, Wang X, Tian JB, Ma PY, Meng FX, Zhang Q, Yu BF, Guo R, Liu ZZ, Wang HL, Xie J, Cheng NL, Wang JH, Niu B, Sun GQ (2019) Construction of a cDNA library and preliminary analysis of the expressed sequence tags of the earthworm Eisenia fetida (Savigny, 1826). Molecular Medicine Reports, 19, 2537-2550. |
| [22] |
Liu CQ, Liu D, Guo Y, Lu TF, Li XC, Zhang MH, Ma JZ, Ma YH, Guan WJ (2013) Construction of a full-length enriched cDNA library and preliminary analysis of expressed sequence tags from Bengal tiger Panthera Tigris Tigris. International Journal of Molecular Sciences, 14, 11072-11083.
DOI URL |
| [23] | Liu HY, Xu N, Zhang QZ, Wang GB, Xu HM, Ruan HH (2020) Characterization of the complete mitochondrial genome of Drawida gisti (Metagynophora, Moniligastridae) and comparison with other Metagynophora species. Genomics, 112, 3056-3064. |
| [24] |
Liu HY, Zhang YF, Xu W, Fang Y, Ruan HH (2021) Characterization of five new earthworm mitogenomes (Oligochaeta: Lumbricidae): Mitochondrial phylogeny of Lumbricidae. Diversity, 13, 580.
DOI URL |
| [25] |
Liu T, Chen XY, Gong X, Lubbers IM, Jiang YY, Feng W, Li XP, Whalen JK, Bonkowski M, Griffiths BS, Hu F, Liu MQ (2019) Earthworms coordinate soil biota to improve multiple ecosystem functions. Current Biology, 29, 3420-3429.
DOI PMID |
| [26] |
Marchán DF, Decaëns T, Domínguez J, Novo M (2022) Perspectives in earthworm molecular phylogeny: Recent advances in Lumbricoidea and standing questions. Diversity, 14, 30.
DOI URL |
| [27] |
Marchán DF, Fernandez R, de Sosa I, Díaz Cosín DJ, Novo M (2017) Pinpointing cryptic borders: Fine-scale phylogeography and genetic landscape analysis of the Hormogaster elisae complex (Oligochaeta, Hormogastridae). Molecular Phylogenetics and Evolution, 112, 185-193.
DOI PMID |
| [28] |
Marchán DF, Fernández R, Domínguez J, Díaz Cosín DJ, Novo M (2020a) Genome-informed integrative taxonomic description of three cryptic species in the earthworm genus Carpetania (Oligochaeta, Hormogastridae). Systematics and Biodiversity, 18, 203-215.
DOI URL |
| [29] |
Marchán DF, Novo M, Sánchez N, Domínguez J, Díaz Cosín DJ, Fernández R (2020b) Local adaptation fuels cryptic speciation in terrestrial annelids. Molecular Phylogenetics and Evolution, 146, 106767.
DOI URL |
| [30] |
Medina-Sauza RM, Álvarez-Jiménez M, Delhal A, Reverchon F, Blouin M, Guerrero-Analco JA, Cerdán CR, Guevara R, Villain L, Barois I (2019) Earthworms building up soil microbiota, a review. Frontiers in Environmental Science, 7, 81.
DOI URL |
| [31] |
Parolini M, Ganzaroli A, Bacenetti J (2020) Earthworm as an alternative protein source in poultry and fish farming: Current applications and future perspectives. Science of the Total Environment, 734, 139460.
DOI URL |
| [32] |
Paul S, Arumugaperumal A, Rathy R, Ponesakki V, Arunachalam P, Sivasubramaniam S (2018) Data on genome annotation and analysis of earthworm Eisenia fetida. Data in Brief, 20, 525-534.
DOI URL |
| [33] |
Pérez-Losada M, Ricoy M, Marshall JC, Domínguez J (2009) Phylogenetic assessment of the earthworm Aporrectodea caliginosa species complex (Oligochaeta: Lumbricidae) based on mitochondrial and nuclear DNA sequences. Molecular Phylogenetics and Evolution, 52, 293-302.
DOI PMID |
| [34] |
Phillips HRP, Guerra CA, Bartz MLC, Briones MJI, Brown G, Crowther TW, Ferlian O, Gongalsky KB, van den Hoogen J, Krebs J, Orgiazzi A, Routh D, Schwarz B, Bach EM, Bennett JM, Brose U, Decaëns T, König-Ries B, Loreau M, Mathieu J, Mulder C, van der Putten WH, Ramirez KS, Rillig MC, Russell D, Rutgers M, Thakur MP, de Vries FT, Wall DH, Wardle DA, Arai M, Ayuke FO, Baker GH, Beauséjour R, Bedano JC, Birkhofer K, Blanchart E, Blossey B, Bolger T, Bradley RL, Callaham MA, Capowiez Y, Caulfield ME, Choi A, Crotty FV, Crumsey JM, Dávalos A, Diaz Cosin DJ, Dominguez A, Duhour AE, van Eekeren N, Emmerling C, Falco LB, Fernández R, Fonte SJ, Fragoso C, Franco ALC, Fugère M, Fusilero AT, Gholami S, Gundale MJ, López MG, Hackenberger DK, Hernández LM, Hishi T, Holdsworth AR, Holmstrup M, Hopfensperger KN, Lwanga EH, Huhta V, Hurisso TT, Iannone BV III, Iordache M, Joschko M, Kaneko N, Kanianska R, Keith AM, Kelly CA, Kernecker ML, Klaminder J, Koné AW, Kooch Y, Kukkonen ST, Lalthanzara H, Lammel DR, Lebedev IM, Li YQ, Jesus Lidon JB, Lincoln NK, Loss SR, Marichal R, Matula R, Moos JH, Moreno G, Morón-Ríos A, Muys B, Neirynck J, Norgrove L, Novo M, Nuutinen V, Nuzzo V, Pansu J, Paudel S, Pérès G, Pérez-Camacho L, Piñeiro R, Ponge JF, Rashid MI, Rebollo S, Rodeiro-Iglesias J, Rodríguez MÁ, Roth AM, Rousseau GX, Rozen A, Sayad E, van Schaik L, Scharenbroch BC, Schirrmann M, Schmidt O, Schröder B, Seeber J, Shashkov MP, Singh J, Smith SM, Steinwandter M, Talavera JA, Trigo D, Tsukamoto J, Vanek SJ, Virto I, Wackett AA, Warren MW, Wehr NH, Whalen JK, Wironen MB, Wolters V, Zenkova IV, Zhang WX, Cameron EK, Eisenhauer N,Mujeeb Rahman P (2019) Global distribution of earthworm diversity. Science, 366, 480-485.
DOI PMID |
| [35] | Pirooznia M, Gong P, Guan X, Inouye LS, Yang K, Perkins EJ, Deng YP (2007) Cloning, analysis and functional annotation of expressed sequence tags from the earthworm Eisenia fetida. BMC Bioinformatics, 8, S7. |
| [36] | Rajesh D, Anant S (2020) Eisenia fetida and Eisenia andrei delimitation by automated barcode gap discovery and neighbor-joining analyses: A review. Journal of Applied Biology & Biotechnology, 8, 93-100. |
| [37] | Seto A, Endo H, Minamiya Y, Matsuda M (2021) The complete mitochondrial genome sequences of Japanese earthworms Metaphire hilgendorfi and Amynthas yunoshimensis (Clitellata: Megascolecidae). Mitochondrial DNA Part B, Resources, 6, 965-967. |
| [38] |
Shao Y, Wang XB, Zhang JJ, Li ML, Wu SS, Ma XY, Wang X, Zhao HF, Li Y, Zhu HH, Irwin DM, Wang DP, Zhang GJ, Ruan J, Wu DD (2020) Genome and single-cell RNA-sequencing of the earthworm Eisenia andrei identifies cellular mechanisms underlying regeneration. Nature Communications, 11, 2656.
DOI URL |
| [39] |
Shekhovtsov SV, Efremov YR, Poluboyarova TV, Peltek SE (2021) Variation in nuclear genome size within the Eisenia nordenskioldi complex (Lumbricidae, Annelida). Vavilov Journal of Genetics and Breeding, 25, 647-651.
DOI URL |
| [40] |
Shekhovtsov SV, Golovanova EV, Ershov NI, Poluboyarova TV, Berman DI, Bulakhova NA, Szederjesi T, Peltek SE (2020) Phylogeny of the Eisenia nordenskioldi complex based on mitochondrial genomes. European Journal of Soil Biology, 96, 103137.
DOI URL |
| [41] |
Shekhovtsov SV, Golovanova EV, Peltek SE (2016) Mitochondrial DNA variation in Eisenia n. nordenskioldi (Lumbricidae) in Europe and Southern Urals. Mitochondrial DNA Part A, 27, 4643-4645.
PMID |
| [42] |
Shekhovtsov SV, Peltek SE (2019) The complete mitochondrial genome of Aporrectodea rosea (Annelida: Lumbricidae). Mitochondrial DNA Part B, 4, 1752-1753.
DOI URL |
| [43] |
Shi ZM, Tang ZW, Wang CY (2017) A brief review and evaluation of earthworm biomarkers in soil pollution assessment. Environmental Science and Pollution Research, 24, 13284-13294.
DOI URL |
| [44] |
Sinha RK, Chauhan K, Valani D, Chandran V, Soni BK, Patel V (2010) Earthworms: Charles Darwin’s ‘unheralded soldiers of mankind’: Protective & productive for man & environment. Journal of Environmental Protection, 1, 251-260.
DOI URL |
| [45] |
Velki M, Ečimović S (2017) Important issues in ecotoxicological investigations using earthworms. Reviews of Environmental Contamination and Toxicology, 239, 157-184.
DOI PMID |
| [46] | Wang AR, Hong Y, Win TM, Kim I (2015) Complete mitochondrial genome of the Burmese giant earthworm, Tonoscolex birmanicus (Clitellata: Megascolecidae). Mitochondrial DNA, 26, 467-468. |
| [47] |
Wang X, Zhang Y, Zhang YF, Kang MM, Li YB, James SW, Yang Y, Bi YM, Jiang H, Zhao Y, Sun ZJ (2021) Amynthas corticis genome reveals molecular mechanisms behind global distribution. Communications Biology, 4, 135.
DOI URL |
| [48] |
Yuan Z, Jiang J, Dong Y, Zhao Q, Sun J, Qiu J (2020) Unearthing the genetic divergence and gene flow of the earthworm Amynthas_YN 2017 sp. (Oligochaeta: Megascolecidae) populations based on restriction site-associated DNA sequencing. European Journal of Soil Biology, 99, 103210.
DOI URL |
| [49] |
Zhang LL, Jiang JB, Dong Y, Qiu JP (2015) Complete mitochondrial genome of four pheretimoid earthworms (Clitellata: Oligochaeta) and their phylogenetic reconstruction. Gene, 574, 308-316.
DOI PMID |
| [50] | Zhang LL, Jiang JB, Dong Y, Qiu JP (2016a) Complete mitochondrial genome of a pheretimoid earthworm Metaphire vulgaris (Oligochaeta: Megascolecidae). Mitochondrial DNA Part A, DNA Mapping, Sequencing, and Analysis, 27, 297-298. |
| [51] | Zhang LL, Jiang JB, Dong Y, Qiu JP (2016b) Complete mitochondrial genome of an Amynthas earthworm, Amynthas aspergillus (Oligochaeta: Megascolecidae). Mitochondrial DNA Part A, DNA Mapping, Sequencing, and Analysis, 27, 1876-1877. |
| [52] |
Zhang LL, Sechi P, Yuan ML, Jiang JB, Dong Y, Qiu JP (2016c) Fifteen new earthworm mitogenomes shed new light on phylogeny within the Pheretima complex. Scientific Reports, 6, 20096.
DOI URL |
| [53] | Zhang QZ, Liu HY, Zhang YF, Ruan HH (2019) The complete mitochondrial genome of Lumbricus rubellus (Oligochaeta, Lumbricidae) and its phylogenetic analysis. Mitochondrial DNA Part B, Resources, 4, 2677-2678. |
| [54] |
Zhang YF, Ganin GN, Atopkin DM, Wu DH (2020) Earthworm Drawida (Moniligastridae) molecular phylogeny and diversity in Far East Russia and Northeast China. The European Zoological Journal, 87, 180-191.
DOI URL |
| [55] |
Zhang YF, Zhang YM, Wu H, Li CS, Aspe NM, Wu DH (2022) Patterns of genetic variation in the Eisenia nordenskioldi complex (Oligochaeta: Lumbricidae) along an elevation gradient in Northern China. Diversity, 14, 35.
DOI URL |
| [56] |
Zhao HF, Fan SH, Aspe NM, Feng LC, Zhang YF (2022) Characterization of 15 earthworm mitogenomes from Northeast China and its phylogenetic implication (Oligochaeta: Lumbricidae, Moniligastridae). Diversity, 14, 714.
DOI URL |
| [57] |
Zwarycz AS, Nossa CW, Putnam NH, Ryan JF (2015) Timing and scope of genomic expansion within Annelida: Evidence from homeoboxes in the genome of the earthworm Eisenia fetida. Genome Biology and Evolution, 8, 271-281.
DOI URL |
| [1] | Xiaoqiang Lu, Shanshan Dong, Yue Ma, Xu Xu, Feng Qiu, Mingyue Zang, Yaqiong Wan, Luanxin Li, Cigang Yu, Yan Liu. Current status, challenges, and prospects of frontier technologies in biodiversity conservation applications [J]. Biodiv Sci, 2025, 33(4): 24440-. |
| [2] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [3] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [4] | Xuemeng Li, Jibao Jiang, Zenglu Zhang, Xiaojing Liu, Yali Wang, Yizhao Wu, Yinsheng Li, Jiangping Qiu, Qi Zhao. Earthworm biodiversity and its influencing factors in Baotianman National Nature Reserve [J]. Biodiv Sci, 2024, 32(4): 23352-. |
| [5] | Zhaoyang Jing, Keguang Cheng, Heng Shu, Yongpeng Ma, Pingli Liu. Whole genome resequencing approach for conservation biology of endangered plants [J]. Biodiv Sci, 2023, 31(5): 22679-. |
| [6] | Jie Tao, Benqiang Li, Jinghua Cheng, Ying Shi, Peihong Liu, Guixia He, Weijie Xu, Huili Liu. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces [J]. Biodiv Sci, 2023, 31(11): 23170-. |
| [7] | Ruiliang Zhu, Xiaoying Ma, Chang Cao, Ziyin Cao. Advances in research on bryophyte diversity in China [J]. Biodiv Sci, 2022, 30(7): 22378-. |
| [8] | Congcong Lu, Qian Liu, Xiaolei Huang. Mitochondrial genome data of three aphid species [J]. Biodiv Sci, 2022, 30(7): 22204-. |
| [9] | Jiman Li, Nan Jin, Maogang Xu, Jusong Huo, Xiaoyun Chen, Feng Hu, Manqiang Liu. Effects of earthworm on tomato resistance under different drought levels [J]. Biodiv Sci, 2022, 30(7): 21488-. |
| [10] | Qi Zhao, Jibao Jiang, Zenglu Zhang, Qing Jin, Jiali Li, Jiangping Qiu. Species composition and phylogenetic analysis of earthworms on Hainan Island [J]. Biodiv Sci, 2022, 30(12): 22224-. |
| [11] | Shanlin Liu, Na Qiu, Shuyi Zhang, Zhunan Zhao, Xin Zhou. Application of genomics technology in biodiversity conservation research [J]. Biodiv Sci, 2022, 30(10): 22441-. |
| [12] | Yuanyuan Li, Chaonan Liu, Rong Wang, Shuixing Luo, Shouqian Nong, Jingwen Wang, Xiaoyong Chen. Applications of molecular markers in conserving endangered species [J]. Biodiv Sci, 2020, 28(3): 367-375. |
| [13] | Wei Wang, Yang Liu. The current status, problems, and policy suggestions for reconstructing the plant tree of life [J]. Biodiv Sci, 2020, 28(2): 176-188. |
| [14] | Xu Yakun, Ma Yue, Hu Xiaoxi, Wang Jun. Analysis of prospective microbiology research using third-generation sequencing technology [J]. Biodiv Sci, 2019, 27(5): 534-542. |
| [15] | Xinyu Du, Jinmei Lu, Dezhu Li. Advances in the evolution of plastid genome structure in lycophytes and ferns [J]. Biodiv Sci, 2019, 27(11): 1172-1183. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()