



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (8): 24501. DOI: 10.17520/biods.2024501 cstr: 32101.14.biods.2024501
• Special Feature: Genetic Diversity and Conservation • Previous Articles Next Articles
Zhicheng Zhou1, Tianling Cao1, Ruyao Liu1, Qiqi Ding1, Ke Ma1, Liping Yang1( ), Chuanjiang Zhou2, Guoxing Nie1(
), Chuanjiang Zhou2, Guoxing Nie1( ), Yongtao Tang1,*(
), Yongtao Tang1,*( )(
)( )
)
Received:2024-11-18
Accepted:2025-04-10
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: tangyongtao@htu.edu.cn
Supported by:Zhicheng Zhou, Tianling Cao, Ruyao Liu, Qiqi Ding, Ke Ma, Liping Yang, Chuanjiang Zhou, Guoxing Nie, Yongtao Tang. Genetic diversity and structure of Pseudorasbora parva population in the Yellow River basin based on the mitochondrial COI gene[J]. Biodiv Sci, 2025, 33(8): 24501.
| 编号 Code | 种群 Population | 采集地点 Sampling location |
|---|---|---|
| 1 | 海东 HD | 青海省海东市互助土族自治县南门峡镇; 南门峡水库 Nanmenxia Town, Huzhu Tu Autonomous County, Haidong City, Qinghai Province; Nanmenxia Reservoir |
| 2 | 兰州 LZ | 甘肃省兰州市河口镇八盘村; 黄河干流 Bapan Village, Hekou Town, Lanzhou City, Gansu Province; mainstream of the Yellow River |
| 3 | 固原 GY | 宁夏回族自治区固原市彭阳县城阳乡刘河村; 茹河 Liuhe Village, Chengyang Town, Pengyang County, Guyuan City, Ningxia Hui Autonomous Region; Ruhe River |
| 4 | 石嘴山 SZS | 宁夏回族自治区石嘴山市惠农区大桥村; 黄河干流 Daqiao Village, Huinong District, Shizuishan City, Ningxia Hui Autonomous Region; mainstream of the Yellow River |
| 5 | 巴彦淖尔 BYNE | 内蒙古自治区巴彦淖尔市乌拉特前旗额尔登布拉格苏木阿日齐嘎查村; 乌良素海 Ariqi Gacha Village, Erden Bulag Sum, Urad Front Banner, Bayannur City, Inner Mongolia Autonomous Region; Ulan Suhai Lake |
| 6 | 吕梁 LL | 山西省吕梁市兴县黑峪口; 黄河干流 Heiyukou, Xingxian County, Lüliang City, Shanxi Province; mainstream of the Yellow River |
| 7 | 太原 TY | 山西省太原市晋源区姚村镇; 汾河 Yaocun Town, Jinyuan District, Taiyuan City, Shanxi Province; Fenhe River |
| 8 | 永和 YH | 山西省临汾市永和县芝河镇红花沟村; 芝河 Honghuagou Village, Zhihe Town, Yonghe County, Linfen City, Shanxi Province; Zhihe River |
| 9 | 大宁 DN | 山西省临汾市大宁县; 昕水河 Daning County, Linfen City, Shanxi Province; Xinshui River |
| 10 | 西安 XA | 陕西省西安市蓝田县孟村镇成中村; 灞河 Chengzhong Village, Mengcun Town, Lantian County, Xi’an City, Shaanxi Province; Bahe River |
| 11 | 洛阳 LY | 河南省洛阳市伊川县韦村; 伊河 Weicun Village, Yichuan County, Luoyang City, Henan Province; Yihe River |
| 12 | 济源 JY | 河南省济源市王屋镇西坪村; 大店河 Xiping Village, Wangwu Town, Jiyuan City, Henan Province; Dadian River |
| 13 | 长垣 CY | 河南省新乡市长垣县芦南乡寨长石桥; 文岩渠 Zhaichangshiqiao, Lunan Town, Changyuan County, Xinxiang City, Henan Province; Wenyan Canal |
| 14 | 泰安 TA | 山东省泰安市大津口乡赤鳞鱼保护区; 天龙水库 Chilinyu Nature Reserve, Dajinkou Town, Tai’an City, Shandong Province; Tianlong Reservoir |
| 15 | 东营 DY | 山东省东营市垦利区西双河村; 黄河干流 Xishuanghe Village, Kenli District, Dongying City, Shandong Province; mainstream of the Yellow River |
Table 1 Sampling information of Pseudorasbora parva in the Yellow River
| 编号 Code | 种群 Population | 采集地点 Sampling location |
|---|---|---|
| 1 | 海东 HD | 青海省海东市互助土族自治县南门峡镇; 南门峡水库 Nanmenxia Town, Huzhu Tu Autonomous County, Haidong City, Qinghai Province; Nanmenxia Reservoir |
| 2 | 兰州 LZ | 甘肃省兰州市河口镇八盘村; 黄河干流 Bapan Village, Hekou Town, Lanzhou City, Gansu Province; mainstream of the Yellow River |
| 3 | 固原 GY | 宁夏回族自治区固原市彭阳县城阳乡刘河村; 茹河 Liuhe Village, Chengyang Town, Pengyang County, Guyuan City, Ningxia Hui Autonomous Region; Ruhe River |
| 4 | 石嘴山 SZS | 宁夏回族自治区石嘴山市惠农区大桥村; 黄河干流 Daqiao Village, Huinong District, Shizuishan City, Ningxia Hui Autonomous Region; mainstream of the Yellow River |
| 5 | 巴彦淖尔 BYNE | 内蒙古自治区巴彦淖尔市乌拉特前旗额尔登布拉格苏木阿日齐嘎查村; 乌良素海 Ariqi Gacha Village, Erden Bulag Sum, Urad Front Banner, Bayannur City, Inner Mongolia Autonomous Region; Ulan Suhai Lake |
| 6 | 吕梁 LL | 山西省吕梁市兴县黑峪口; 黄河干流 Heiyukou, Xingxian County, Lüliang City, Shanxi Province; mainstream of the Yellow River |
| 7 | 太原 TY | 山西省太原市晋源区姚村镇; 汾河 Yaocun Town, Jinyuan District, Taiyuan City, Shanxi Province; Fenhe River |
| 8 | 永和 YH | 山西省临汾市永和县芝河镇红花沟村; 芝河 Honghuagou Village, Zhihe Town, Yonghe County, Linfen City, Shanxi Province; Zhihe River |
| 9 | 大宁 DN | 山西省临汾市大宁县; 昕水河 Daning County, Linfen City, Shanxi Province; Xinshui River |
| 10 | 西安 XA | 陕西省西安市蓝田县孟村镇成中村; 灞河 Chengzhong Village, Mengcun Town, Lantian County, Xi’an City, Shaanxi Province; Bahe River |
| 11 | 洛阳 LY | 河南省洛阳市伊川县韦村; 伊河 Weicun Village, Yichuan County, Luoyang City, Henan Province; Yihe River |
| 12 | 济源 JY | 河南省济源市王屋镇西坪村; 大店河 Xiping Village, Wangwu Town, Jiyuan City, Henan Province; Dadian River |
| 13 | 长垣 CY | 河南省新乡市长垣县芦南乡寨长石桥; 文岩渠 Zhaichangshiqiao, Lunan Town, Changyuan County, Xinxiang City, Henan Province; Wenyan Canal |
| 14 | 泰安 TA | 山东省泰安市大津口乡赤鳞鱼保护区; 天龙水库 Chilinyu Nature Reserve, Dajinkou Town, Tai’an City, Shandong Province; Tianlong Reservoir |
| 15 | 东营 DY | 山东省东营市垦利区西双河村; 黄河干流 Xishuanghe Village, Kenli District, Dongying City, Shandong Province; mainstream of the Yellow River |
| 河段/流域 Reach/Basin | 种群 Population | 样本数 Sample size | 多态位点 Polymorphic loci | 单倍型数 No. of haplotypes | 单倍型分布 Haplotype distribution |
|---|---|---|---|---|---|
| 黄河上游河段 Upper reaches of the Yellow River | 海东 HD | 10 | 11 | 6 | Hap 3, Hap 4, Hap 5, Hap 7, Hap 8, Hap 24 |
| 兰州 LZ | 9 | 14 | 7 | Hap 1, Hap 2, Hap 3, Hap 4, Hap 11, Hap 12, Hap 13 | |
| 固原 GY | 14 | 9 | 4 | Hap 1, Hap 5, Hap 7, Hap 16 | |
| 石嘴山 SZS | 11 | 6 | 3 | Hap 1, Hap 3, Hap 7 | |
| 巴彦淖尔 BYNE | 14 | 1 | 2 | Hap 7, Hap 14 | |
| 整体 Overall | 58 | 20 | 13 | Hap 1, Hap 2, Hap 3, Hap 4, Hap 5, Hap 7, Hap 8, Hap 11, Hap 12, Hap 13, Hap 14, Hap 16, Hap 24 | |
| 黄河中游河段 Middle reaches of the Yellow River | 吕梁 LL | 13 | 11 | 7 | Hap 1, Hap 3, Hap 4, Hap 5, Hap 6, Hap 7, Hap 17 |
| 太原 TY | 7 | 11 | 6 | Hap 3, Hap 7, Hap 8, Hap 16, Hap 22, Hap 23 | |
| 永和 YH | 7 | 8 | 3 | Hap 3, Hap 7, Hap 22 | |
| 大宁 DN | 22 | 13 | 9 | Hap 1, Hap 3, Hap 4, Hap 7, Hap 8, Hap 15, Hap 19, Hap 20, Hap 21 | |
| 西安 XA | 8 | 12 | 7 | Hap 1, Hap 3, Hap 5, Hap 6, Hap 7, Hap 15, Hap 18 | |
| 洛阳 LY | 8 | 11 | 6 | Hap 1, Hap 3, Hap 5, Hap 7, Hap 8, Hap 9 | |
| 济源 JY | 5 | 6 | 3 | Hap 3, Hap 7, Hap 8 | |
| 整体 Overall | 70 | 23 | 17 | Hap 1, Hap 3, Hap 4, Hap 5, Hap 6, Hap 7, Hap 8, Hap 9, Hap 15, Hap 16, Hap 17, Hap 18, Hap 19, Hap 20, Hap 21, Hap 22, Hap 23 | |
| 黄河下游河段 Lower reaches of the Yellow River | 长垣 CY | 5 | 9 | 4 | Hap 1, Hap 4, Hap 5, Hap 10 |
| 泰安 TA | 10 | 1 | 2 | Hap 7, Hap 23 | |
| 东营 DY | 10 | 7 | 7 | Hap 7, Hap 8, Hap 19, Hap 23, Hap 25, Hap 26, Hap 27 | |
| 整体 Overall | 25 | 15 | 11 | Hap 1, Hap 4, Hap 5, Hap 7, Hap 8, Hap 10, Hap 19, Hap 23, Hap 25, Hap 26, Hap 27 | |
| 黄河 Yellow River | 总计 Total | 153 | 34 | 27 | Hap 1-Hap 27 |
| 雅鲁藏布江 Yarlung Zangbo River | 雅鲁藏布江 YLZBJ | 1 | - | 1 | Hap 3 |
| 珠江 Pearl River | 珠江 ZJ | 18 | 2 | 3 | Hap 4, Hap 28, Hap 29 |
| 长江 Yangtze River | 长江 CJ | 21 | 13 | 8 | Hap 1, Hap 4, Hap 6, Hap 7, Hap 30, Hap 31, Hap 32, Hap 33 |
| 怒江 Nujiang River | 怒江 NJ | 54 | 4 | 5 | Hap 1, Hap 3, Hap 4, Hap 28, Hap 34 |
| 海河 Haihe River | 海河 HaiHe | 10 | 10 | 6 | Hap 1, Hap 5, Hap 6, Hap 7, Hap 8, Hap 23 |
| 淮河 Huaihe River | 淮河 HuaiHe | 10 | 10 | 6 | Hap 4, Hap 5, Hap 7, Hap 8, Hap 35, Hap 36 |
| 鸭绿江 Yalu River | 鸭绿江 YLJ | 2 | - | 1 | Hap 37 |
Table 2 Haplotype information of COI sequences of Pseudorasbora parva populations in the Yellow River basin and other river systems
| 河段/流域 Reach/Basin | 种群 Population | 样本数 Sample size | 多态位点 Polymorphic loci | 单倍型数 No. of haplotypes | 单倍型分布 Haplotype distribution |
|---|---|---|---|---|---|
| 黄河上游河段 Upper reaches of the Yellow River | 海东 HD | 10 | 11 | 6 | Hap 3, Hap 4, Hap 5, Hap 7, Hap 8, Hap 24 |
| 兰州 LZ | 9 | 14 | 7 | Hap 1, Hap 2, Hap 3, Hap 4, Hap 11, Hap 12, Hap 13 | |
| 固原 GY | 14 | 9 | 4 | Hap 1, Hap 5, Hap 7, Hap 16 | |
| 石嘴山 SZS | 11 | 6 | 3 | Hap 1, Hap 3, Hap 7 | |
| 巴彦淖尔 BYNE | 14 | 1 | 2 | Hap 7, Hap 14 | |
| 整体 Overall | 58 | 20 | 13 | Hap 1, Hap 2, Hap 3, Hap 4, Hap 5, Hap 7, Hap 8, Hap 11, Hap 12, Hap 13, Hap 14, Hap 16, Hap 24 | |
| 黄河中游河段 Middle reaches of the Yellow River | 吕梁 LL | 13 | 11 | 7 | Hap 1, Hap 3, Hap 4, Hap 5, Hap 6, Hap 7, Hap 17 |
| 太原 TY | 7 | 11 | 6 | Hap 3, Hap 7, Hap 8, Hap 16, Hap 22, Hap 23 | |
| 永和 YH | 7 | 8 | 3 | Hap 3, Hap 7, Hap 22 | |
| 大宁 DN | 22 | 13 | 9 | Hap 1, Hap 3, Hap 4, Hap 7, Hap 8, Hap 15, Hap 19, Hap 20, Hap 21 | |
| 西安 XA | 8 | 12 | 7 | Hap 1, Hap 3, Hap 5, Hap 6, Hap 7, Hap 15, Hap 18 | |
| 洛阳 LY | 8 | 11 | 6 | Hap 1, Hap 3, Hap 5, Hap 7, Hap 8, Hap 9 | |
| 济源 JY | 5 | 6 | 3 | Hap 3, Hap 7, Hap 8 | |
| 整体 Overall | 70 | 23 | 17 | Hap 1, Hap 3, Hap 4, Hap 5, Hap 6, Hap 7, Hap 8, Hap 9, Hap 15, Hap 16, Hap 17, Hap 18, Hap 19, Hap 20, Hap 21, Hap 22, Hap 23 | |
| 黄河下游河段 Lower reaches of the Yellow River | 长垣 CY | 5 | 9 | 4 | Hap 1, Hap 4, Hap 5, Hap 10 |
| 泰安 TA | 10 | 1 | 2 | Hap 7, Hap 23 | |
| 东营 DY | 10 | 7 | 7 | Hap 7, Hap 8, Hap 19, Hap 23, Hap 25, Hap 26, Hap 27 | |
| 整体 Overall | 25 | 15 | 11 | Hap 1, Hap 4, Hap 5, Hap 7, Hap 8, Hap 10, Hap 19, Hap 23, Hap 25, Hap 26, Hap 27 | |
| 黄河 Yellow River | 总计 Total | 153 | 34 | 27 | Hap 1-Hap 27 |
| 雅鲁藏布江 Yarlung Zangbo River | 雅鲁藏布江 YLZBJ | 1 | - | 1 | Hap 3 |
| 珠江 Pearl River | 珠江 ZJ | 18 | 2 | 3 | Hap 4, Hap 28, Hap 29 |
| 长江 Yangtze River | 长江 CJ | 21 | 13 | 8 | Hap 1, Hap 4, Hap 6, Hap 7, Hap 30, Hap 31, Hap 32, Hap 33 |
| 怒江 Nujiang River | 怒江 NJ | 54 | 4 | 5 | Hap 1, Hap 3, Hap 4, Hap 28, Hap 34 |
| 海河 Haihe River | 海河 HaiHe | 10 | 10 | 6 | Hap 1, Hap 5, Hap 6, Hap 7, Hap 8, Hap 23 |
| 淮河 Huaihe River | 淮河 HuaiHe | 10 | 10 | 6 | Hap 4, Hap 5, Hap 7, Hap 8, Hap 35, Hap 36 |
| 鸭绿江 Yalu River | 鸭绿江 YLJ | 2 | - | 1 | Hap 37 |
| 河段 Reach | 种群 Population | 单倍型多样性 Haplotype diversity (h) | 核苷酸多样性 Nucleotide diversity (π) | 平均核苷酸差异数 Average number of nucleotide differences (k) | |||||
|---|---|---|---|---|---|---|---|---|---|
| 上游河段 Upper reaches | 海东 HD | 0.844 ± 0.103 | 0.00549 ± 0.00113 | 3.422 | |||||
| 兰州 LZ | 0.944 ± 0.070 | 0.00687 ± 0.00141 | 4.278 | ||||||
| 固原 GY | 0.626 ± 0.110 | 0.00349 ± 0.00101 | 2.176 | ||||||
| 石嘴山 SZS | 0.473 ± 0.162 | 0.00362 ± 0.00115 | 2.255 | ||||||
| 巴彦淖尔 BYNE | 0.143 ± 0.119 | 0.00023 ± 0.00019 | 0.143 | ||||||
| 整体 Overall | 0.665 ± 0.068 | 0.00445 ± 0.00058 | 2.771 | ||||||
| 中游河段 Middle reaches | 吕梁 LL | 0.833 ± 0.086 | 0.00539 ± 0.00082 | 3.359 | |||||
| 太原 TY | 0.952 ± 0.096 | 0.00627 ± 0.00178 | 3.905 | ||||||
| 永和 YH | 0.762 ± 0.115 | 0.00657 ± 0.00116 | 4.095 | ||||||
| 大宁 DN | 0.818 ± 0.072 | 0.00482 ± 0.00083 | 3.004 | ||||||
| 西安 XA | 0.964 ± 0.077 | 0.00682 ± 0.00125 | 4.250 | ||||||
| 洛阳 LY | 0.929 ± 0.084 | 0.00659 ± 0.00134 | 4.107 | ||||||
| 济源 JY | 0.800 ± 0.164 | 0.00417 ± 0.00189 | 2.600 | ||||||
| 整体 Overall | 0.863 ± 0.029 | 0.00579 ± 0.00046 | 3.607 | ||||||
| 下游河段 Lower reaches | 长垣 CY | 0.900 ± 0.161 | 0.00803 ± 0.00213 | 5.000 | |||||
| 泰安 TA | 0.467 ± 0.132 | 0.00075 ± 0.00021 | 0.467 | ||||||
| 东营 DY | 0.911 ± 0.077 | 0.00342 ± 0.00067 | 2.133 | ||||||
| 整体 Overall | 0.827 ± 0.059 | 0.00397 ± 0.00079 | 2.473 | ||||||
| 总计 Total | 0.802 ± 0.030 | 0.00508 ± 0.00035 | 3.162 | ||||||
| 其他水系 Other rivers | 珠江 ZJ | 0.582 ± 0.061 | 0.00103 ± 0.00017 | 0.641 | |||||
| 长江 CJ | 0.767 ± 0.068 | 0.00368 ± 0.00097 | 2.295 | ||||||
| 怒江 NJ | 0.715 ± 0.031 | 0.00178 ± 0.00014 | 1.109 | ||||||
| 海河 HaiHe | 0.911 ± 0.062 | 0.00564 ± 0.00564 | 3.511 | ||||||
| 淮河 HuaiHe | 0.867 ± 0.085 | 0.00471 ± 0.00117 | 2.933 |
Table 3 Genetic diversity of Pseudorasbora parva populations in the Yellow River basin and other river systems
| 河段 Reach | 种群 Population | 单倍型多样性 Haplotype diversity (h) | 核苷酸多样性 Nucleotide diversity (π) | 平均核苷酸差异数 Average number of nucleotide differences (k) | |||||
|---|---|---|---|---|---|---|---|---|---|
| 上游河段 Upper reaches | 海东 HD | 0.844 ± 0.103 | 0.00549 ± 0.00113 | 3.422 | |||||
| 兰州 LZ | 0.944 ± 0.070 | 0.00687 ± 0.00141 | 4.278 | ||||||
| 固原 GY | 0.626 ± 0.110 | 0.00349 ± 0.00101 | 2.176 | ||||||
| 石嘴山 SZS | 0.473 ± 0.162 | 0.00362 ± 0.00115 | 2.255 | ||||||
| 巴彦淖尔 BYNE | 0.143 ± 0.119 | 0.00023 ± 0.00019 | 0.143 | ||||||
| 整体 Overall | 0.665 ± 0.068 | 0.00445 ± 0.00058 | 2.771 | ||||||
| 中游河段 Middle reaches | 吕梁 LL | 0.833 ± 0.086 | 0.00539 ± 0.00082 | 3.359 | |||||
| 太原 TY | 0.952 ± 0.096 | 0.00627 ± 0.00178 | 3.905 | ||||||
| 永和 YH | 0.762 ± 0.115 | 0.00657 ± 0.00116 | 4.095 | ||||||
| 大宁 DN | 0.818 ± 0.072 | 0.00482 ± 0.00083 | 3.004 | ||||||
| 西安 XA | 0.964 ± 0.077 | 0.00682 ± 0.00125 | 4.250 | ||||||
| 洛阳 LY | 0.929 ± 0.084 | 0.00659 ± 0.00134 | 4.107 | ||||||
| 济源 JY | 0.800 ± 0.164 | 0.00417 ± 0.00189 | 2.600 | ||||||
| 整体 Overall | 0.863 ± 0.029 | 0.00579 ± 0.00046 | 3.607 | ||||||
| 下游河段 Lower reaches | 长垣 CY | 0.900 ± 0.161 | 0.00803 ± 0.00213 | 5.000 | |||||
| 泰安 TA | 0.467 ± 0.132 | 0.00075 ± 0.00021 | 0.467 | ||||||
| 东营 DY | 0.911 ± 0.077 | 0.00342 ± 0.00067 | 2.133 | ||||||
| 整体 Overall | 0.827 ± 0.059 | 0.00397 ± 0.00079 | 2.473 | ||||||
| 总计 Total | 0.802 ± 0.030 | 0.00508 ± 0.00035 | 3.162 | ||||||
| 其他水系 Other rivers | 珠江 ZJ | 0.582 ± 0.061 | 0.00103 ± 0.00017 | 0.641 | |||||
| 长江 CJ | 0.767 ± 0.068 | 0.00368 ± 0.00097 | 2.295 | ||||||
| 怒江 NJ | 0.715 ± 0.031 | 0.00178 ± 0.00014 | 1.109 | ||||||
| 海河 HaiHe | 0.911 ± 0.062 | 0.00564 ± 0.00564 | 3.511 | ||||||
| 淮河 HuaiHe | 0.867 ± 0.085 | 0.00471 ± 0.00117 | 2.933 |
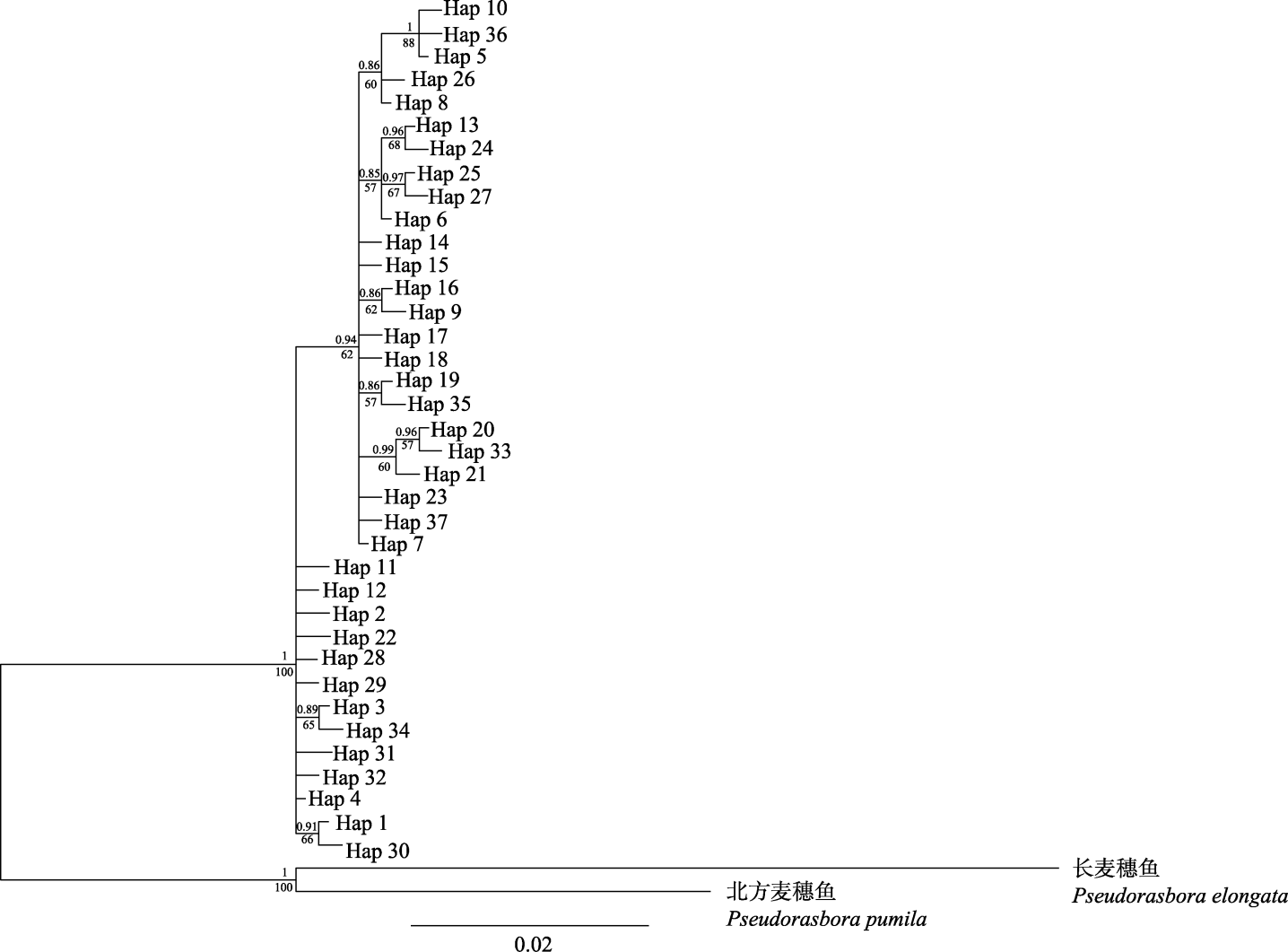
Fig. 2 Bayesian inference (BI) and maximum likelihood (ML) tree of Pseudorasbora parva based on COI sequences. Values at the nodes represent support values from BI and ML analyses respectively.
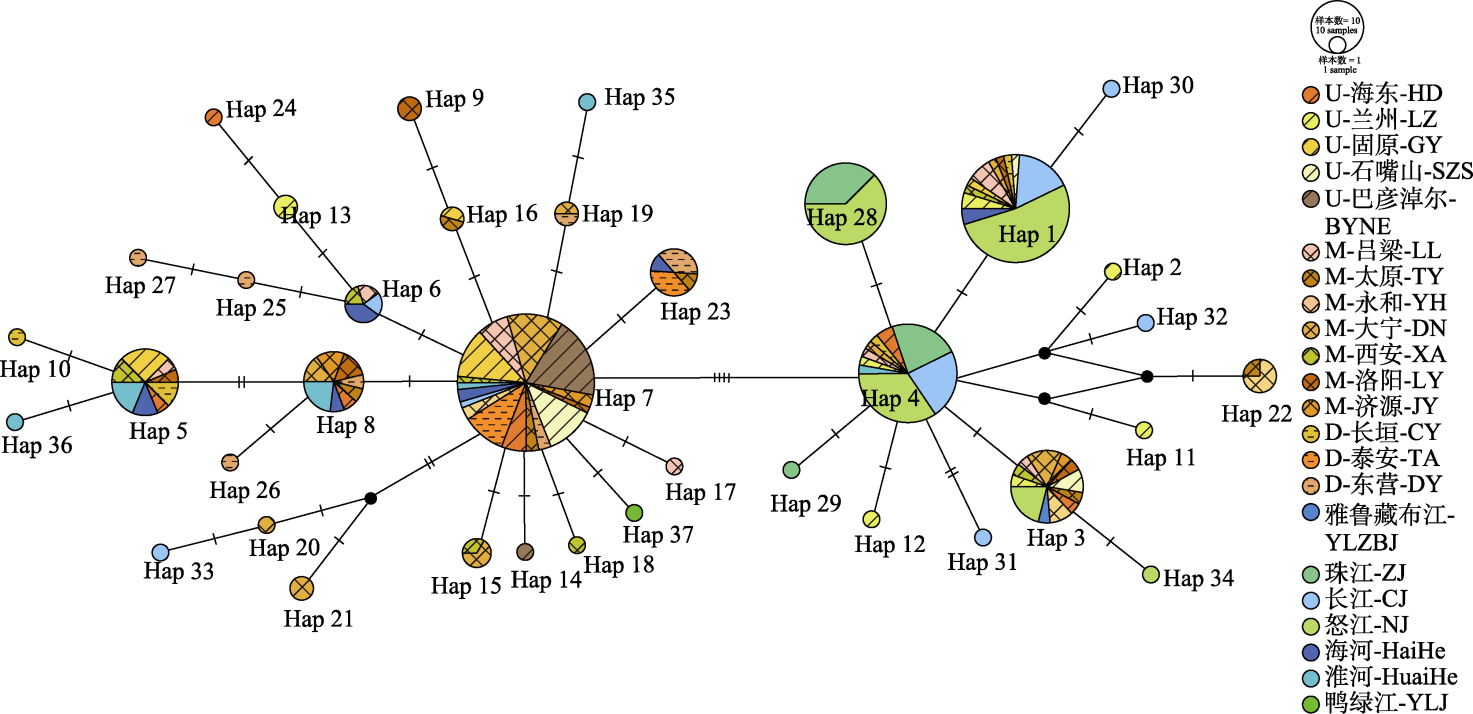
Fig. 3 The haplotype network of Pseudorasbora parva based on the Templeton-Crandall-Sing (TCS) model. Meanings of the sampling site abbreviations are provided in Table 1.
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of squares | 变异组成 Variance components | 变异百分比 Percentage of variation | 遗传分化系数 Fixation index |
|---|---|---|---|---|---|
| 组间 Among groups | 2 | 8.783 | 0.00205 | 0.11% | 0.00112 |
| 组内种群间 Among populations within groups | 12 | 48.398 | 0.24715 | 13.51% | 0.13528** |
| 种群内 Within populations | 138 | 218.011 | 1.57979 | 86.38% | 0.13625** |
| 总计 Total | 152 | 275.191 | 1.82899 | 100% |
Table 4 AMOVA analysis based on the COI sequences of Pseudorasbora parva from the Yellow River basin
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of squares | 变异组成 Variance components | 变异百分比 Percentage of variation | 遗传分化系数 Fixation index |
|---|---|---|---|---|---|
| 组间 Among groups | 2 | 8.783 | 0.00205 | 0.11% | 0.00112 |
| 组内种群间 Among populations within groups | 12 | 48.398 | 0.24715 | 13.51% | 0.13528** |
| 种群内 Within populations | 138 | 218.011 | 1.57979 | 86.38% | 0.13625** |
| 总计 Total | 152 | 275.191 | 1.82899 | 100% |
| 河段 Reach | 种群 Population | Tajima’s D | Fu’s Fs | 偏差平方和 Sum of squared deviations test (SSD) | 粗糙指数 Harpending’s raggedness index (Hri) |
|---|---|---|---|---|---|
| 上游河段 Upper reaches | 海东 HD | -0.5368 | -0.5021 | 0.0180 | 0.0558 |
| 兰州 LZ | -0.8138 | -1.6069 | 0.0324 | 0.0741 | |
| 固原 GY | -0.8872 | 1.4138 | 0.0687 | 0.3195 | |
| 石嘴山 SZS | 0.3982 | 2.4788 | 0.1328 | 0.6615 | |
| 巴彦淖尔 BYNE | -1.1552 | -0.5948 | 0.0001 | 0.5306 | |
| 整体 Overall | -1.1903 | -1.5391 | 0.0616 | 0.1692 | |
| 中游河段 Middle reaches | 吕梁 LL | -0.2118 | -0.7978 | 0.0451 | 0.0988 |
| 太原 TY | -0.7048 | -1.6423 | 0.0584 | 0.1293 | |
| 永和 YH | 1.3352 | 2.9764 | 0.0907 | 0.3469 | |
| 大宁 DN | -0.5580 | -1.4627 | 0.0207 | 0.0524 | |
| 西安 XA | -0.4111 | -2.3041 | 0.0420 | 0.1594 | |
| 洛阳 LY | -0.1593 | -0.9017 | 0.0323 | 0.0740 | |
| 济源 JY | -0.6682 | 1.0900 | 0.1222 | 0.2800 | |
| 整体 Overall | -0.7548 | -3.3261 | 0.0153 | 0.0256 | |
| 下游河段 Lower reaches | 长垣 CY | 1.1186 | 0.4897 | 0.1303 | 0.2300 |
| 泰安 TA | 0.8198 | 0.8180 | 0.0158** | 0.2222 | |
| 东营 DY | -0.5838 | -3.0656* | 0.0049 | 0.0573 | |
| 整体 Overall | -1.3214 | -3.8449* | 0.0100 | 0.0355 | |
| 总计 Total | -1.3881 | -9.70418* | 0.0099 | 0.0279 |
Table 5 Neutrality tests for all populations of Pseudorasbora parva from the Yellow River basin
| 河段 Reach | 种群 Population | Tajima’s D | Fu’s Fs | 偏差平方和 Sum of squared deviations test (SSD) | 粗糙指数 Harpending’s raggedness index (Hri) |
|---|---|---|---|---|---|
| 上游河段 Upper reaches | 海东 HD | -0.5368 | -0.5021 | 0.0180 | 0.0558 |
| 兰州 LZ | -0.8138 | -1.6069 | 0.0324 | 0.0741 | |
| 固原 GY | -0.8872 | 1.4138 | 0.0687 | 0.3195 | |
| 石嘴山 SZS | 0.3982 | 2.4788 | 0.1328 | 0.6615 | |
| 巴彦淖尔 BYNE | -1.1552 | -0.5948 | 0.0001 | 0.5306 | |
| 整体 Overall | -1.1903 | -1.5391 | 0.0616 | 0.1692 | |
| 中游河段 Middle reaches | 吕梁 LL | -0.2118 | -0.7978 | 0.0451 | 0.0988 |
| 太原 TY | -0.7048 | -1.6423 | 0.0584 | 0.1293 | |
| 永和 YH | 1.3352 | 2.9764 | 0.0907 | 0.3469 | |
| 大宁 DN | -0.5580 | -1.4627 | 0.0207 | 0.0524 | |
| 西安 XA | -0.4111 | -2.3041 | 0.0420 | 0.1594 | |
| 洛阳 LY | -0.1593 | -0.9017 | 0.0323 | 0.0740 | |
| 济源 JY | -0.6682 | 1.0900 | 0.1222 | 0.2800 | |
| 整体 Overall | -0.7548 | -3.3261 | 0.0153 | 0.0256 | |
| 下游河段 Lower reaches | 长垣 CY | 1.1186 | 0.4897 | 0.1303 | 0.2300 |
| 泰安 TA | 0.8198 | 0.8180 | 0.0158** | 0.2222 | |
| 东营 DY | -0.5838 | -3.0656* | 0.0049 | 0.0573 | |
| 整体 Overall | -1.3214 | -3.8449* | 0.0100 | 0.0355 | |
| 总计 Total | -1.3881 | -9.70418* | 0.0099 | 0.0279 |
| [1] |
Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research, 25, 4692-4693.
DOI PMID |
| [2] |
Baltazar-Soares M, Blanchet S, Cote J, Tarkan AS, Záhorská E, Gozlan RE, Eizaguirre C (2020) Genomic footprints of a biological invasion: Introduction from Asia and dispersal in Europe of the topmouth gudgeon (Pseudorasbora parva). Molecular Ecology, 29, 71-85.
DOI PMID |
| [3] | Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 10, e1003537. |
| [4] | Chen YY (1998) Fauna Sinica·Osteichthyes·Cypriniformes (II). Science Press, Beijing. (in Chinese) |
| [陈宜瑜 (1998) 中国动物志·硬骨鱼纲·鲤形目(中卷). 科学出版社, 北京.] | |
| [5] |
Davison PI, Créach V, Liang WJ, Andreou D, Britton JR, Copp GH (2016) Laboratory and field validation of a simple method for detecting four species of non-native freshwater fish using eDNA. Journal of Fish Biology, 89, 1782-1793.
DOI PMID |
| [6] | Ding HP (2019) The Comparative Ecology of Pseudorasbora parva from Different Regions in China. PhD dissertation, Chinese Academy of Sciences, Beijing. (in Chinese with English abstract) |
| [丁慧萍 (2019) 中国不同地理域麦穗鱼种群的比较生态学研究. 博士学位论文, 中国科学院大学, 北京.] | |
| [7] |
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [8] |
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915-925.
DOI PMID |
| [9] | Gao JX, Yu D, Liu HZ (2021) Genetic diversity and phylogeography of Opsariichthys macrolepis from the upper and middle Yangtze River based on Cytochrome b sequences. Sichuan Journal of Zoology, 40, 361-373. (in Chinese with English abstract) |
| [高嘉昕, 俞丹, 刘焕章 (2021) 基于Cyt b基因的长江中上游大鳞马口鱼遗传多样性及谱系生物地理学过程分析. 四川动物, 40, 361-373.] | |
| [10] |
Grant W, Bowen B (1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426.
DOI URL |
| [11] |
He M, Fang DA, Chen YJ, Sun HB, Luo H, Ren YF, Li TY (2023) Genetic diversity evaluation and conservation of Topmouth Culter (Culter alburnus) germplasm in five river basins in China. Biology, 12, 12.
DOI URL |
| [12] | He YF, Zhu YJ, Gong JL, Zhu TB, Wu XB, Li XM, Meng ZH, Yang DG (2022) Genetic diversity and population demography of Coreius guichenoti from the middle and lower reaches of the Jinsha River. Acta Hydrobiologica Sinica, 46, 37-47. (in Chinese with English abstract) |
| [何勇凤, 朱永久, 龚进玲, 朱挺兵, 吴兴兵, 李学梅, 孟子豪, 杨德国 (2022) 金沙江中下游圆口铜鱼遗传多样性与种群历史动态分析. 水生生物学报, 46, 37-47.] | |
| [13] |
Jia YT, Kennard MJ, Liu YH, Sui XY, Chen YY, Li KM, Wang GJ, Chen YF (2019) Understanding invasion success of Pseudorasbora parva in the Qinghai-Tibetan Plateau: Insights from life-history and environmental filters. Science of the Total Environment, 694, 133739.
DOI URL |
| [14] |
Klütsch CFC, Maduna SN, Polikarpova NV, Forfang K, Aspholm PE, Nyman T, Eiken HG, Amundsen PA, Hagen SB (2019) Genetic changes caused by restocking and hydroelectric dams in demographically bottlenecked brown trout in a transnational subarctic riverine system. Ecology and Evolution, 9, 6068-6081.
DOI URL |
| [15] |
Leigh JW, Bryant D (2015) Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110-1116.
DOI URL |
| [16] | Liang HY (2018) Phylogeography of Tamarix austromongolica and T. chinensis, Endemic Tree Species Distributed along the Yellow River. PhD dissertation, Henan Agricultural University, Zhengzhou. (in Chinese with English abstract) |
| [梁红艳 (2018) 黄河流域特有种甘蒙柽柳和柽柳的谱系地理学研究. 博士学位论文, 河南农业大学, 郑州.] | |
| [17] |
Liu DQ, Lan F, Xie SC, Diao Y, Zheng Y, Gong JH (2021) Dynamic genetic diversity and population structure of Coreius guichenoti. ZooKeys, 1055, 135-148.
DOI URL |
| [18] | Ma HY, Guo JF, Yue YS (2006) Genetic variation of Pseudorasbora parva in Dongping Lake using microsatellites. Life Science Research, 10(1), 67-70. (in Chinese with English abstract) |
| [马洪雨, 郭金峰, 岳永生 (2006) 东平湖麦穗鱼群体遗传结构的微卫星标记分析. 生命科学研究, 10(1), 67-70.] | |
| [19] | Ministry of Water Resources of the People’s Republic of China (2024) China Water Statistical Yearbook 2024. China Water & Power Press, Beijing. (in Chinese) |
| [中华人民共和国水利部 (2024) 中国水利统计年鉴(2024). 中国水利水电出版社, 北京.] | |
| [20] | Oksanen J, Blanchet FG, Friendly M, Kindt R (2008) R package for Community Ecologists: Popular Ordination Methods, Ecological Null Models & Diversity Analysis. https://github.com/vegandevs/vegan . (accessed on 2024-03-12) |
| [21] | Qinghai Provincial Local Chronicles Compilation Committee (1993) Qinghai Provincial Chronicles (Vol. 12): Agriculture and Fisheries. Qinghai People’s Publishing House, Xining. (in Chinese) |
| [青海省地方志编纂委员会 (1993) 青海省志(第12卷): 农业志, 渔业志. 青海人民出版社, 西宁.] | |
| [22] | Rambaut A (2012) FigTree v1.4.3. http://tree.bio.ed.ac.uk/software/figtree/ . (accessed on 2024-02-21) |
| [23] |
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67, 901-904.
DOI PMID |
| [24] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299-3302.
DOI PMID |
| [25] |
Ruzich J, Turnquist K, Nye N, Rowe D, Larson WA (2019) Isolation by a hydroelectric dam induces minimal impacts on genetic diversity and population structure in six fish species. Conservation Genetics, 20, 1421-1436.
DOI |
| [26] |
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution, 47, 264-279.
DOI PMID |
| [27] | Sun HH, Zhu R, Feng X, Xiong W, Sui XY, Chen YF (2022) Adaptation in life-history traits of exotic topmouth gudgeon from wetlands in Tibet Autonomous Region. Acta Hydrobiologica Sinica, 46, 1780-1787. (in Chinese with English abstract) |
| [孙欢欢, 朱仁, 冯秀, 熊文, 隋晓云, 陈毅峰 (2022) 西藏湿地外来麦穗鱼生活史特征的适应性研究. 水生生物学报, 46, 1780-1787.] | |
| [28] |
Swindell SR, Plasterer TN (1997) SEQMAN. Contig assembly. Methods in Molecular Biology, 70, 75-89.
PMID |
| [29] |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
DOI PMID |
| [30] |
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38, 3022-3027.
DOI PMID |
| [31] |
Tang WJ, He DK (2015) Investigation on alien fishes in Qinghai Province, China (2001-2014). Journal of Lake Sciences, 27, 502-510. (in Chinese with English abstract)
DOI URL |
| [唐文家, 何德奎 (2015) 青海省外来鱼类调查(2001-2014年). 湖泊科学, 27, 502-510.] | |
| [32] |
Wang YH, Tan D, Han LY, Li DH, Wang X, Lu GY, Lin JJ (2021) Review of climate change in the Yellow River Basin. Journal of Desert Research, 41(4), 235-246. (in Chinese with English abstract)
DOI |
|
[王有恒, 谭丹, 韩兰英, 李丹华, 王鑫, 卢国阳, 林婧婧 (2021) 黄河流域气候变化研究综述. 中国沙漠, 41(4), 235-246.]
DOI |
|
| [33] |
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences, 360, 1847-1857.
DOI URL |
| [34] |
Wu YF, Wu CZ (1990) A preliminary study of the fishery resources and the countermeasures of fishery development in the region of the Karakorum-Kunlun mountains, China. Journal of Natural Resources, 5, 354-364. (in Chinese with English abstract)
DOI |
| [武云飞, 吴翠珍 (1990) 喀喇昆仑山-昆仑山地区渔业资源及渔业发展对策的初步研究. 自然资源学报, 5, 354-364.] | |
| [35] | Wu YF, Wu CZ (1992) The Fishes of the Qinghai-Xizang Plateau. Sichuan Science and Technology Press, Chengdu. (in Chinese) |
| [武云飞, 吴翠珍 (1992) 青藏高原鱼类. 四川科学技术出版社, 成都.] | |
| [36] | Xu RG (2013) Fauna of Inner Mongolia (Vol. 1). Mongolia University Press, Hohhot. (in Chinese) |
| [旭日干 (2013) 内蒙古动物志(第一卷). 内蒙古大学出版社, 呼和浩特.] | |
| [37] | Yang JQ (2005) Molecular Phylogeny, Evolutionary Process and Biogeography of the Gobioninae (Pisces:Cyprinidae). PhD dissertation, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan. (in Chinese with English abstract) |
| [杨金权 (2005) 鮈亚科鱼类分子系统发育、演化过程及生物地理学研究. 博士学位论文, 中国科学院水生生物研究所, 武汉.] | |
| [38] | Yang LP, Hu JY, Qin CB, Zhang YR, Lu RH, Meng XL, Yang GK, Yan X, Zhi SY, Nie GX (2020) Genetic structure analysis of Pseudorasbora parva in the four major river systems in Yunnan based on mitochondrial Cyt b. Journal of Fisheries of China, 44, 339-350. (in Chinese with English abstract) |
| [杨丽萍, 胡俊仪, 秦超彬, 张玉茹, 卢荣华, 孟晓林, 杨国坤, 闫潇, 职韶阳, 聂国兴 (2020) 基于线粒体Cyt b分析入侵云南四大水系的麦穗鱼群体遗传结构. 水产学报, 44, 339-350.] | |
| [39] |
Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348-355.
DOI PMID |
| [40] |
Zhao YH, Xing YC, Lü BB, Zhou CJ, Yang WB, Zhao K (2020) Species diversity and conservation of freshwater fishes in the Yellow River basin. Biodiversity Science, 28, 1496-1510. (in Chinese with English abstract)
DOI URL |
| [赵亚辉, 邢迎春, 吕彬彬, 周传江, 杨文波, 赵凯 (2020) 黄河流域淡水鱼类多样性和保护. 生物多样性, 28, 1496-1510.] | |
| [41] | Zuo DQ (2001) Historical evolution of Yellow River channel pattern and its enlightenment to ideal of present Yellow River regulation. Advances in Science and Technology of Water Resources, 21(4), 2-13, 69. (in Chinese with English abstract) |
| [左东启 (2001) 黄河河道格局的历史演变及其对现代治黄思路的启示. 水利水电科技进展, 21(4), 2-13, 69.] |
| [1] | Ping Fan, Zhixin Wen, Gang Song. The effect of climatic factors and anthropogenic activities on different genetic diversity indicators of amphibians and mammals [J]. Biodiv Sci, 2025, 33(8): 25022-. |
| [2] | Ruixiang Xue, Xuerong Ma, Jiongwen Wu, Aijun Liu, Xiquan Zhang, Congliang Ji, Yingshan Yin, Weijian Zhu, Qingbin Luo. Genetic diversity and genetic structure of Zhongshan partridge duck populations [J]. Biodiv Sci, 2025, 33(8): 24592-. |
| [3] | Ping Fan, Huan Wang, Zhixin Wen, Gang Song, Fuming Lei. Impact of climatic factors on the genetic diversity-species area relationship of birds [J]. Biodiv Sci, 2025, 33(8): 25072-. |
| [4] | Qilong Yu, Minhui Hao, Huaijiang He, Chunyu Zhang, Xiuhai Zhao. Relationships of biodiversity and productivity change with forest succession in Changbai Mountains: Insights from species, traits, and phylogeny [J]. Biodiv Sci, 2025, 33(8): 25060-. |
| [5] | Ruxiao Wang, Boyang Shi, Da Pan, Hongying Sun. Biodiversity patterns and conservation gaps of the endemic freshwater crab genus Sinopotamon in China [J]. Biodiv Sci, 2025, 33(8): 25123-. |
| [6] | Yongqiang Shi, Xiujuan Shan, Jie Zhao, Yinuo Wang, Qingshan Luan, Xiaodong Bian, Yunlong Chen, Xianshi Jin. Evolution of zooplankton taxa composition and biodiversity in the Yellow River estuary and its adjacent waters from 1958 to 2020 [J]. Biodiv Sci, 2025, 33(7): 24437-. |
| [7] | Zhang Mingyi, Wang Xiaomei, Zheng Yanxin, Wu Nan, Li Donghao, Fan Enyuan, Li Na, Shan Xiujuan, Yu Tao, Zhao Chunnuan, Li Bo, Xu Shuai, Wu Yuping, Ren Liqun. Resource status and habitat function of typical oyster reef areas in the Yellow River Estuary [J]. Biodiv Sci, 2025, 33(4): 24208-. |
| [8] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [9] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [10] | Yanmei Ni, Li Chen, Zhiyuan Dong, Debin Sun, Baoquan Li, Xumin Wang, Linlin Chen. Community structure of macrobenthos and ecological health evaluation in the restoration area of the Yellow River Delta wetland [J]. Biodiv Sci, 2024, 32(3): 23303-. |
| [11] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [12] | Chunxiao Wang, Zhengwang Zhang, Shaoxia Xia, Houlang Duan, Wen Wang, Yifei Jia, Lixun Zhang, Gang Feng, Yaqiao Yang, Tong Li, Changqing Ding, Chunping Wang, Baodong Yuan, Jinyu Lei, Yu Liu, Jianbin Shi, Keqi Lan, Qingqing Shi, Qing Xiao, Xiubo Yu. Seasonal and regional patterns and conservation strategies of waterbird diversity in the Yellow River Basin [J]. Biodiv Sci, 2024, 32(11): 23490-. |
| [13] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [14] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [15] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()