



Biodiv Sci ›› 2021, Vol. 29 ›› Issue (3): 409-418. DOI: 10.17520/biods.2020276 cstr: 32101.14.biods.2020276
Special Issue: 物种形成与系统进化
• Forum • Previous Articles
Minlan Li1,2, Chao Wang1,*( ), Ruiwu Wang1,*(
), Ruiwu Wang1,*( )
)
Received:2020-07-11
Accepted:2021-01-04
Online:2021-03-20
Published:2021-01-13
Contact:
Chao Wang,Ruiwu Wang
About author:First author contact:#Co-first authors
Minlan Li, Chao Wang, Ruiwu Wang. Path-dependent speciation in the process of evolution[J]. Biodiv Sci, 2021, 29(3): 409-418.
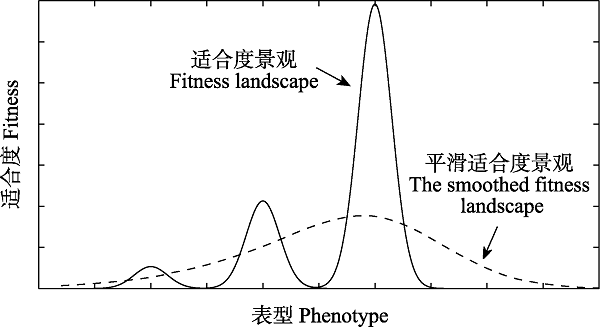
Fig. 1 Fitness landscape of phenotypes. The solid line shows the fitness landscape observed. When an organism evolves on such a multipeak fitness landscape, it’s easy to be trapped at a lower local peak. The dashed line displays the phenotypic fitness landscape smoothed by reaction norm, i.e., the expect fitness for a genotype with a certain average phenotype. This fitness landscape just has one single peak, which makes it easier for any phenotype to evolve to another one with higher fitness.
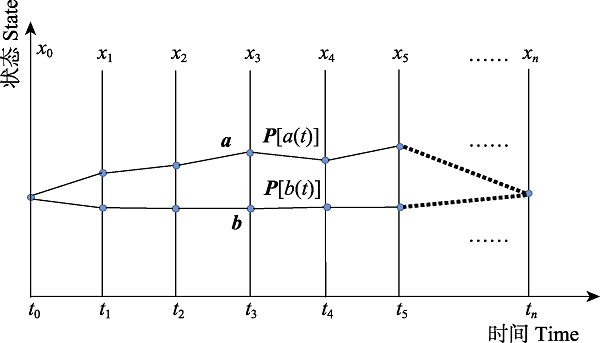
Fig. 2 An evolutionary process for phenotype in a population from x0 to xn. An individual selects the next phenotype to evolve with a certain probability according to its instantaneous fitness at each time step, thus many possible paths of phenotype in a population occur including paths a and b. Suppose that trajectory a consists of phenotype ${{x}_{0}},{{x}_{1}},{{x}_{2}},...,{{x}_{n}}$, then the possibility is $P[{{x}_{0}}({{t}_{0}}),{{x}_{1}}({{t}_{1}}),{{x}_{2}}({{t}_{2}}),\ldots,{{x}_{n}}({{t}_{n}})]$。
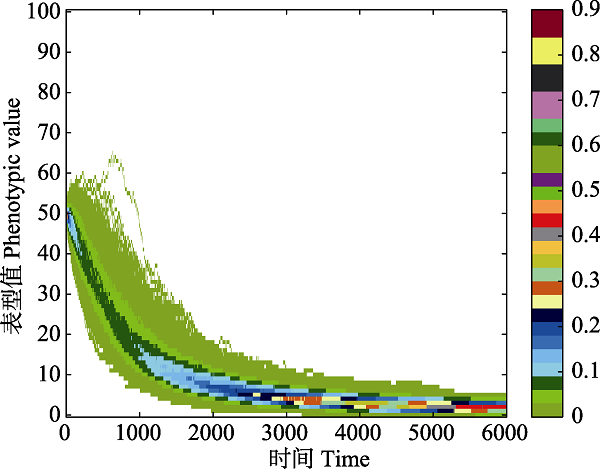
Fig. 3 The distribution of evolutionary paths. Different colors represent different distribution probability of paths. As time goes by, one phenotype differentiates to other phenotypes with different probabilities. The evolutionary path is continuous in this process because that phenotypes can only evolve to adjacent phenotypes. If the evolutionary results are observed at the moment of t = 1,000, significant discontinuous phenotype differentiation (blue regions) can be seen.
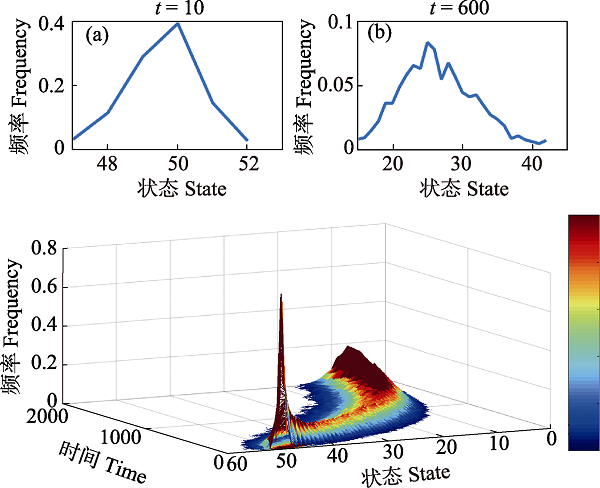
Fig. 4 Path-dependent evolutionary process of states. (a) The frequency distribution curve of states over time. The states represent genotype, phenotype or ecological process. (b) The frequency distribution curve of phenotypes at t = 10, (c) The frequency distribution curve of phenotypes at t = 600. From the perspective of phenotype, the distribution curve changes from unimodal in the early phase to bimodal at t = 600, which means the phenotype differentiates into two phenotypes (25 and 28) at t = 600 from one phenotype (50) in the initial stage. Then we can judge if the species with phenotype 25 and 28 are different species.
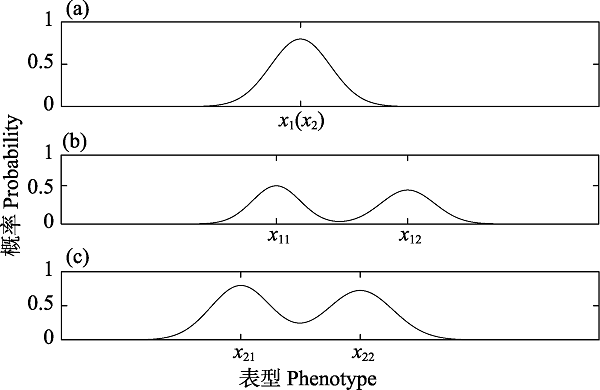
Fig. 5 Frequency distribution of phenotypes and genotypes at different time. (a) showing the frequency distribution curve of the state x1(x2) in the early differentiation. (b) demonstrating the frequency distribution curve of the phenotype x1 at a certain moment after differentiation. (c) indicating the frequency distribution curve of the genotype x2 at the same moment after differentiation. Suppose that any two states x1 and x2 of the original species A evolved influenced by random mutation and drift, then different degree of differentiation occurs in the same or different time. When observing in a “cutting plane” of a species evolution, we notice that x1(x2) differentiates into x11(x21) and x12(x22), and the evolution are path-dependent. Meanwhile, if the differentiated phenotypes and the genotypes in a quantity statistics satisfy certain conditions, the individuals with ${{x}_{11}},{{x}_{21}}({{x}_{11}},{{x}_{22}},or{{x}_{12}},{{x}_{21}},or{{x}_{12}},{{x}_{22}})$ are new species, respectively.
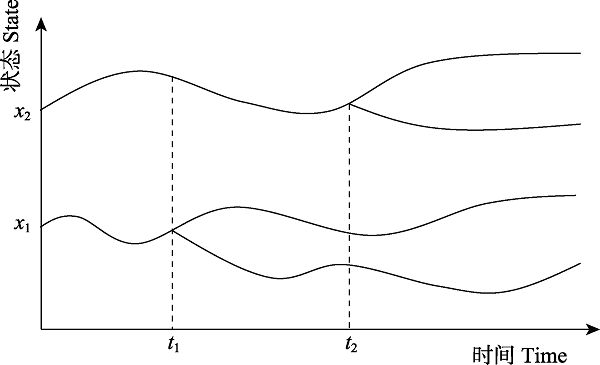
Fig. 6 schematic diagram of the species delimitation over time. The state x1 (x2) differentiates at t1(t2). It can be determined whether a specie at different moments is a new species.
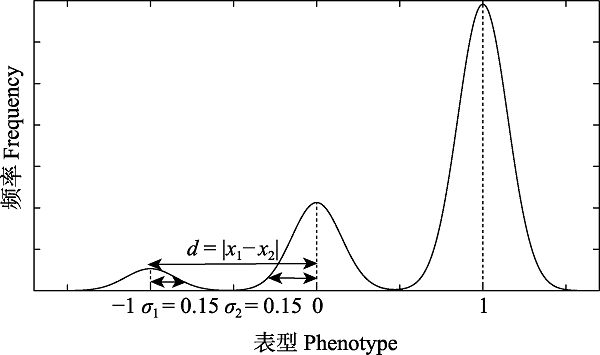
Fig. 7 Differentiation of three independent phenotypes from one phenotype of a species at a moment. The figure is a special case of judging whether the phenotypic differentiates completely. There is a continuous and a little difference for the phenotype ?1 within the species (the difference range is ± 0.15), and the phenotype 0 versa. The discontinuity of these two interspecies phenotypes is 1, and the discontinuity is much higher than the intraspecies continuous difference. Therefore, it can be considered that the phenotype meets the requirements for phenotype in the concept of morphological species. Taking into account the differentiation of another state of this species, if the difference satisfies the same conditions, the corresponding individuals can be determined as a new species.
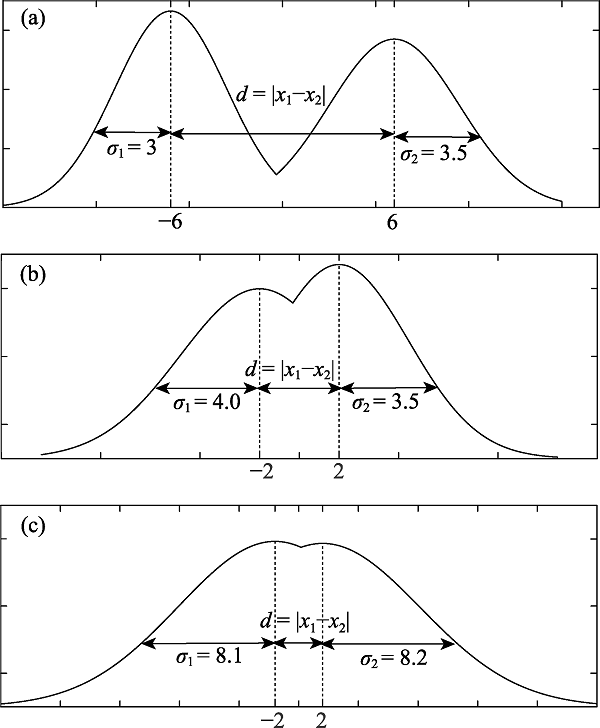
Fig. 8 Different case of phenotype differentiation. Imagine the relation of difference has been satisfied for one biological character. Then we consider phenotypes. In figure 8(a), $d=|{{x}_{1}}-{{x}_{2}}|=|(-6)-6|=12,{{\sigma }_{1}}=3,{{\sigma }_{2}}=3.5,d>{{\sigma }_{1}}$ and $d>{{\sigma }_{2}}$. If individuals in two populations satisfy these conditions, they are different species. In figure 8(b), $d=|{{x}_{1}}-{{x}_{2}}|=|(-2)-2|=4,{{\sigma }_{1}}=4,{{\sigma }_{2}}=3.5,d={{\sigma }_{1}}$ and $d>{{\sigma }_{2}}$. If those individuals satisfy these conditions, they are the same species. In figure 8(c), $d=|{{x}_{1}}-{{x}_{2}}|=|(-2)-2|=4,{{\sigma }_{1}}=8.1,{{\sigma }_{2}}=8.2,d<{{\sigma }_{1}}$ and $d<{{\sigma }_{2}}$. If those individuals satisfy these conditions, they are the same species.
| [1] |
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JBW, Zinner D (2013) Hybridization and speciation. Journal of Evolutionary Biology, 26,229-246.
URL PMID |
| [2] | Darwin C (1929) The origin of species by means of natural selection. American Anthropologist, 61,176-177. |
| [3] |
Frank SA (2011) Natural selection. II. Developmental variability and evolutionary rate. Journal of Evolutionary Biology, 24,2310-2320.
URL PMID |
| [4] | Hedberg O (1958) The taxonomic treatment of vicarious taxa. Uppsala Universitets Arsskrift, 6,186-195. |
| [5] | Hong DY (2016) Biodiversity pursuits need a scientific and operative species concept. Biodiversity Science, 24,979-999. (in Chinese with English abstract) |
| [ 洪德元 (2016) 生物多样性事业需要科学、可操作的物种概念. 生物多样性, 24,979-999.]. | |
| [6] | Li QJ, Li Y (2010) Lamarck redux—A revisit of Darwinism. Journal of Biology, 27(2),55-57. (in Chinese with English abstract) |
| [ 李启剑, 李越 (2010) 拉马克的归来: 对达尔文主义的再审视. 生物学杂志, 27(2),55-57.]. | |
| [7] | Liu JQ (2016) “The integrative species concept” and “species on the speciation way”. Biodiversity Science, 24,1004-1008. (in Chinese with English abstract) |
| [ 刘建全 (2016) “整合物种概念”和“分化路上的物种”. 生物多样性, 24,1004-1008.]. | |
| [8] | Lu BR, Wang Z (2016) What is a species: Conflict between evolutionary continuity and taxonomic discontinuity. Chinese Science Bulletin, 61,2663-2669. (in Chinese with English abstract) |
| [ 卢宝荣, 王哲 (2016) 什么是物种: 进化连续性与分类间断性冲突的产物. 科学通报, 61,2663-2669.]. | |
| [9] | Maynard Smith J (1982) Evolution and the theory of games. Cambridge University Press, Cambridge. |
| [10] | Traulsen A, Iwasa Y, Nowak MA (2007) The fastest evolutionary trajectory. Journal of Theoretical Biology, 249,617-623. |
| [11] | Nowak MA (2006) Evolutionary Dynamics:Exploring the Equations of Life. Harvard University Press, Cambridge. |
| [12] | Wang RW (2021) The End of Rationality and Selfness—A Story on the Asymmetry, Uncertainty and Evolution of Cooperation. China Commerce and Trade Press, Beijing. (in Chinese) (in press) |
| 王瑞武 (2021) 理性与自私的终结——非对称性、不确定性与社会合作行为. 中国商务出版社, 北京. | |
| [13] | Wilkins JS (2009) Species: A History of the Idea. University of California Press, Berkeley. |
| [14] | Zhou CF, Yang G (2011) Existence and Definition of Species. Science Press, Beijing. (in Chinese) |
| 周长发, 杨光 (2011) 物种的存在与定义. 科学出版社, 北京. |
| [1] | Xiao-Qing Wu Meihui Zhang Suting Ge Manshu Li Kun Song Guochun Shen Jian Zhang. Spatiotemporal Dynamics of Woody Plant Species Diversity and Aboveground Biomass during Near-Natural Forest Reconstruction in Shanghai: A Case Study from the Eco-Island in Minhang District [J]. Biodiv Sci, 2025, 33(5): 24444-. |
| [2] | Murong Yi, Ping Lu, Yong Peng, Yong Tang, Jiuheng Xu, Haoping Yin, Luyang Zhang, Xiaodong Weng, Mingxiao Di, Juan Lei, Chenqi Lu, Rujun Cao, Nianhua Dai, Deyang Zhan, Mei Tong, Zhiming Lou, Yonggang Ding, Jing Chai, Jing Che. Population status and habitat of Critically Endangered Jiangxi giant salamander (Andrias jiangxiensis) [J]. Biodiv Sci, 2025, 33(4): 24145-. |
| [3] | Gan Xie, Jing Xuan, Qidi Fu, Ze Wei, Kai Xue, Hairui Luo, Jixi Gao, Min Li. Establishing an intelligent identification model for unmanned aerial vehicle surveys of grassland plant diversity [J]. Biodiv Sci, 2025, 33(4): 24236-. |
| [4] | Wei Tian, Jiajie Chen, Yuange Chen, Qing Xu, Jin Zhou. Species diversity of crabs (Decapoda: Branchyura) of Xiangshan Bay, Zhejiang Province [J]. Biodiv Sci, 2025, 33(4): 24461-. |
| [5] | Tai Wang, Fujun Song, Yongsheng Zhang, Zhongyu Lou, Yanping Zhang, Yanyan Du. Fish diversity and resource status in interior drainage systems of Hexi Corridor [J]. Biodiv Sci, 2025, 33(4): 24387-. |
| [6] | Motong Li, Tuo He, Wei Li, Jing Liao, Yan Zeng. Regulating international trade in wild fauna and flora: An analysis of CITES terminology [J]. Biodiv Sci, 2025, 33(4): 24545-. |
| [7] | Jingjing Zhang, Wenbin Huang, Yiting Chen, Zepeng Yang, Weiye Ke, Zhaojie Peng, Shichao Wei, Zhiwei Zhang, Yisi Hu, Wenhua Yu, Wenliang Zhou. Reef-building coral diversity and distribution characteristics in the National Nature Reserve for Marine Ecology of Guangdong Nanpeng Islands [J]. Biodiv Sci, 2025, 33(4): 24424-. |
| [8] | Jingyi Yuan, Xu Zhang, Zhenpeng Tian, Zizhe Wang, Yongping Gao, Dizhao Yao, Hongcan Guan, Wenkai Li, Jing Liu, Hong Zhang, Qin Ma. A comparison of methods for extracting tree species composition and quantitative characteristics in urban plant communities using UAV high-resolution RGB imagery and LiDAR point cloud [J]. Biodiv Sci, 2025, 33(4): 24237-. |
| [9] | Guo Yutong, Li Sucui, Wang Zhi, Xie Yan, Yang Xue, Zhou Guangjin, You Chunhe, Zhu Saning, Gao Jixi. Coverage and distribution of national key protected wild species in China’s nature reserves [J]. Biodiv Sci, 2025, 33(3): 24423-. |
| [10] | Jia Zhenni, Zhang Yicen, Du Yanjun, Ren Haibao. Influences of disturbances on successional dynamics of species diversity in mid- subtropical forests [J]. Biodiv Sci, 2025, 33(2): 24078-. |
| [11] | Ma Wenjun, Liu Sijia, Li Kemao, Jian Shenglong, Xue Chang’an, Han Qingxiango, Wei Jinliang, Chen Shengxue, Niu Yimeng, Cui Zhouping, Sui Ruichen, Tian Fei, Zhao Kai. Fish diversity and distribution in the source region of the Yangtze River in Qinghai Province [J]. Biodiv Sci, 2025, 33(2): 24494-. |
| [12] | Shang Huadan, Zhang Chuqing, Wang Mei, Pei Wenya, Li Guohong, Wang Hongbin. Species diversity and geographic distribution of poplar pests in China [J]. Biodiv Sci, 2025, 33(2): 24370-. |
| [13] | Wu Yuxuan, Wang Ping, Hu Xiaosheng, Ding Yi, Peng Tiantian, Zhi Qiuying, Bademu Qiqige, Li Wenjie, Guan Xiao, Li Junsheng. Evaluation of grassland degradation status and vegetation characteristics changes in Hulunbuir [J]. Biodiv Sci, 2025, 33(2): 24118-. |
| [14] | Wang Dawei, Cheng Shuai, Feng Jiawei, Wang Tianming. The wildlife camera-trapping dataset of Zhangguangcai Mountains in Northeast China (2015-2020) [J]. Biodiv Sci, 2025, 33(2): 24384-. |
| [15] | Hong Deyuan. A brief discussion on methodology in taxonomy [J]. Biodiv Sci, 2025, 33(2): 24541-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()