



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (4): 398-407. DOI: 10.3724/SP.J.1003.2010.398 cstr: 32101.14.SP.J.1003.2010.398
Special Issue: 生物入侵
• Special Issue • Previous Articles Next Articles
Lingliang Cao, Lizhi Zhou( ), Baowei Zhang
), Baowei Zhang
Received:2010-03-02
Accepted:2010-06-13
Online:2010-07-20
Published:2010-07-20
Contact:
Lizhi Zhou
Lingliang Cao, Lizhi Zhou, Baowei Zhang. Genetic patterns of an invasive Procambarus clarkii population in the three river basins of Anhui Province[J]. Biodiv Sci, 2010, 18(4): 398-407.
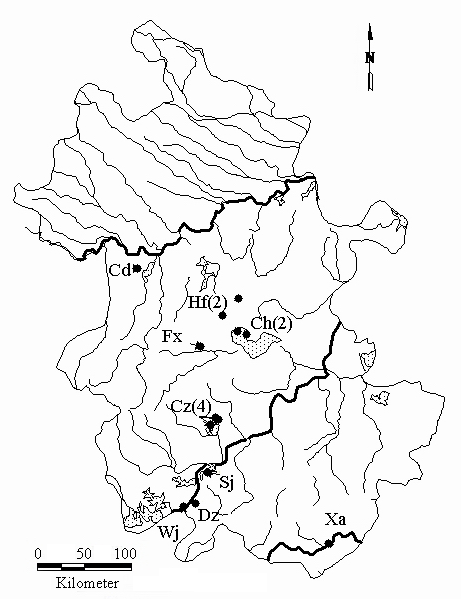
Fig. 1 Geographical distribution of the nine Procambarus clarkii populations in the study. The abbreviated codes refer to local populations in Table 1. The numbers in parentheses indicate sampling times.
| 地方种群 Local populations | 编号 Code | 采集点 Sampling site | 样本量 Sample size | 期望杂合度 HE | 观测杂合度 HO | 平均等位 基因数 MNA | 私有等位基因数 Np |
|---|---|---|---|---|---|---|---|
| 淮河 Huaihe River | |||||||
| 城东湖 Chengdong Lake | Cd | 霍邱黄泊渡 Huangbodu, Huoqiu | 24 | 0.72 | 0.31 | 6.62 | 5 |
| 江淮之间 Between Yangtze River and Huaihe River | |||||||
| 巢湖 Chaohu Lake | Ch | 义城 Yicheng | 7 | 0.65 | 0.24 | 4.89 | 5 |
| 长临河口 Changlin River Mouths | 3 | 0 | |||||
| 肥西 Feixi | Fx | 柿树岗 Stream in Shishugang | 12 | 0.56 | 0.39 | 4.44 | 3 |
| 合肥 Hefei | Hf | 董铺水库 Dongpu Reservoir | 6 | 0.57 | 0.33 | 4.75 | 2 |
| 三十头支流 Stream in Sanshitou | 9 | 1 | |||||
| 长江 Yangtze River | |||||||
| 菜子湖 Caizi Lake | Czh | 王湾咀 Wangwanzui | 7 | 0.67 | 0.47 | 9.89 | 4 |
| 杨湾 Yangwan | 3 | 0 | |||||
| 肖店 Xiaodian | 21 | 6 | |||||
| 双店 Shuangdian | 12 | 3 | |||||
| 升金湖 Shengjin Lake | Sj | 升金湖 Shenjin Lake | 13 | 0.71 | 0.56 | 6.33 | 5 |
| 东至 Dongzhi | Dz | 香口乡 Stream in Xiangkou | 21 | 0.74 | 0.33 | 7.55 | 9 |
| 望江 Wangjiang | Wj | 华阳河 Huayang River | 20 | 0.76 | 0.17 | 7.44 | 18 |
| 新安江 Xin'anjiang River | |||||||
| 新安江 Xin'anjiang River | Xa | 屯光镇 Tunguang | 24 | 0.64 | 0.39 | 6.00 | 3 |
| 总计 Total | 182 | 0.67 | 0.35 | - | - | ||
Table 1 Sample size, study sites and genetic diversity of the Procambarus clarkii populations
| 地方种群 Local populations | 编号 Code | 采集点 Sampling site | 样本量 Sample size | 期望杂合度 HE | 观测杂合度 HO | 平均等位 基因数 MNA | 私有等位基因数 Np |
|---|---|---|---|---|---|---|---|
| 淮河 Huaihe River | |||||||
| 城东湖 Chengdong Lake | Cd | 霍邱黄泊渡 Huangbodu, Huoqiu | 24 | 0.72 | 0.31 | 6.62 | 5 |
| 江淮之间 Between Yangtze River and Huaihe River | |||||||
| 巢湖 Chaohu Lake | Ch | 义城 Yicheng | 7 | 0.65 | 0.24 | 4.89 | 5 |
| 长临河口 Changlin River Mouths | 3 | 0 | |||||
| 肥西 Feixi | Fx | 柿树岗 Stream in Shishugang | 12 | 0.56 | 0.39 | 4.44 | 3 |
| 合肥 Hefei | Hf | 董铺水库 Dongpu Reservoir | 6 | 0.57 | 0.33 | 4.75 | 2 |
| 三十头支流 Stream in Sanshitou | 9 | 1 | |||||
| 长江 Yangtze River | |||||||
| 菜子湖 Caizi Lake | Czh | 王湾咀 Wangwanzui | 7 | 0.67 | 0.47 | 9.89 | 4 |
| 杨湾 Yangwan | 3 | 0 | |||||
| 肖店 Xiaodian | 21 | 6 | |||||
| 双店 Shuangdian | 12 | 3 | |||||
| 升金湖 Shengjin Lake | Sj | 升金湖 Shenjin Lake | 13 | 0.71 | 0.56 | 6.33 | 5 |
| 东至 Dongzhi | Dz | 香口乡 Stream in Xiangkou | 21 | 0.74 | 0.33 | 7.55 | 9 |
| 望江 Wangjiang | Wj | 华阳河 Huayang River | 20 | 0.76 | 0.17 | 7.44 | 18 |
| 新安江 Xin'anjiang River | |||||||
| 新安江 Xin'anjiang River | Xa | 屯光镇 Tunguang | 24 | 0.64 | 0.39 | 6.00 | 3 |
| 总计 Total | 182 | 0.67 | 0.35 | - | - | ||
| 位点 Loci | 重复单元Repeat unit | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature (°C) | 长度 Size range (bp) | 登录号 Accession no. | Na | HE | HO | PIC | Fis | Fit | Fst |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PclG33 | (GT)21 | F: FAM-TTCGAGGCGTTGCTGATTGTAAGT R: CAAGGAAGCGTATAGCCGGAGTCT | 56 | 120-180 | AF290936 | 23 | 0.83 | 0.46 | 0.81 | 0.50 | 0.56 | 0.110 |
| PCL29 | (GA)27 | F: HEX-CACTCAAGCCTGCCCTCACTC R: GTCTCTTCCTCCCCCATTCTCAC | 56 | 170-214 | EF564126 | 14 | 0.87 | 0.44 | 0.85 | 0.55 | 0.55 | 0.108 |
| PCL47 | (TG)13 | F: TAMRA-ACTCTGCCCATTGTTTCTCGG R: AGCCCTTGGACCCCGCCTATC | 56 | 324-344 | EF564128 | 10 | 0.79 | 0.82 | 0.75 | 0.57 | 0.62 | 0.110 |
| PCL33 | (TG)30 | F: FAM-ACTCCTGTCCCATTTCACTAC R: ACAACTAACTGCAACTCATTCTA | 56 | 132-158 | EF564127 | 24 | 0.81 | 0.43 | 0.78 | 0.49 | 0.54 | 0.100 |
| PCL28 | (CA)29 | F: HEX-CCTACCAGAGAACCCAAAACAGAA R: GTCAGCCTCCACCACATCACTT | 56 | 229-239 | EF564125 | 23 | 0.82 | 0.35 | 0.80 | 0.50 | 0.56 | 0.111 |
| PCL17 | (TC)17 | F: TAMRA-TTCACTACCGCCCACAGGATG R: TAGGCACCGCACTTATTAGACCAG | 56 | 370-422 | EF564122 | 18 | 0.71 | 0.10 | 0.69 | 0.45 | 0.51 | 0.111 |
| PclG48 | (CA)12 | F: FAM-CTGTTGGTGATTTCCGTCAATTTT R: AGATTCAACGCTGTGTTCCTGATC | 56 | 146-190 | AF290941 | 22 | 0.81 | 0.24 | 0.79 | 0.47 | 0.53 | 0.108 |
| PCL02 | (TG)30 | F: HEX-GAAGACGGGACACCACGAG R: ATCAAATCAAACGAAGCAAGAAAG | 56 | 245-271 | EF564119 | 24 | 0.86 | 0.23 | 0.84 | 0.46 | 0.52 | 0.113 |
| PCL50 | (GA)12 | F: TAMRA-AAGCGCTGAAATGCACAAACAAGA R: CAAGCCCCGAGGTCAAAGGTC | 56 | 424-525 | EF564129 | 19 | 0.53 | 0.17 | 0.51 | 0.48 | 0.53 | 0.105 |
Table 2 Primer sequence, genetic differentiation, polymorphism information content and Wright’ F-statistics at nine microsatellite loci of Procambarus clarkii
| 位点 Loci | 重复单元Repeat unit | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature (°C) | 长度 Size range (bp) | 登录号 Accession no. | Na | HE | HO | PIC | Fis | Fit | Fst |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PclG33 | (GT)21 | F: FAM-TTCGAGGCGTTGCTGATTGTAAGT R: CAAGGAAGCGTATAGCCGGAGTCT | 56 | 120-180 | AF290936 | 23 | 0.83 | 0.46 | 0.81 | 0.50 | 0.56 | 0.110 |
| PCL29 | (GA)27 | F: HEX-CACTCAAGCCTGCCCTCACTC R: GTCTCTTCCTCCCCCATTCTCAC | 56 | 170-214 | EF564126 | 14 | 0.87 | 0.44 | 0.85 | 0.55 | 0.55 | 0.108 |
| PCL47 | (TG)13 | F: TAMRA-ACTCTGCCCATTGTTTCTCGG R: AGCCCTTGGACCCCGCCTATC | 56 | 324-344 | EF564128 | 10 | 0.79 | 0.82 | 0.75 | 0.57 | 0.62 | 0.110 |
| PCL33 | (TG)30 | F: FAM-ACTCCTGTCCCATTTCACTAC R: ACAACTAACTGCAACTCATTCTA | 56 | 132-158 | EF564127 | 24 | 0.81 | 0.43 | 0.78 | 0.49 | 0.54 | 0.100 |
| PCL28 | (CA)29 | F: HEX-CCTACCAGAGAACCCAAAACAGAA R: GTCAGCCTCCACCACATCACTT | 56 | 229-239 | EF564125 | 23 | 0.82 | 0.35 | 0.80 | 0.50 | 0.56 | 0.111 |
| PCL17 | (TC)17 | F: TAMRA-TTCACTACCGCCCACAGGATG R: TAGGCACCGCACTTATTAGACCAG | 56 | 370-422 | EF564122 | 18 | 0.71 | 0.10 | 0.69 | 0.45 | 0.51 | 0.111 |
| PclG48 | (CA)12 | F: FAM-CTGTTGGTGATTTCCGTCAATTTT R: AGATTCAACGCTGTGTTCCTGATC | 56 | 146-190 | AF290941 | 22 | 0.81 | 0.24 | 0.79 | 0.47 | 0.53 | 0.108 |
| PCL02 | (TG)30 | F: HEX-GAAGACGGGACACCACGAG R: ATCAAATCAAACGAAGCAAGAAAG | 56 | 245-271 | EF564119 | 24 | 0.86 | 0.23 | 0.84 | 0.46 | 0.52 | 0.113 |
| PCL50 | (GA)12 | F: TAMRA-AAGCGCTGAAATGCACAAACAAGA R: CAAGCCCCGAGGTCAAAGGTC | 56 | 424-525 | EF564129 | 19 | 0.53 | 0.17 | 0.51 | 0.48 | 0.53 | 0.105 |
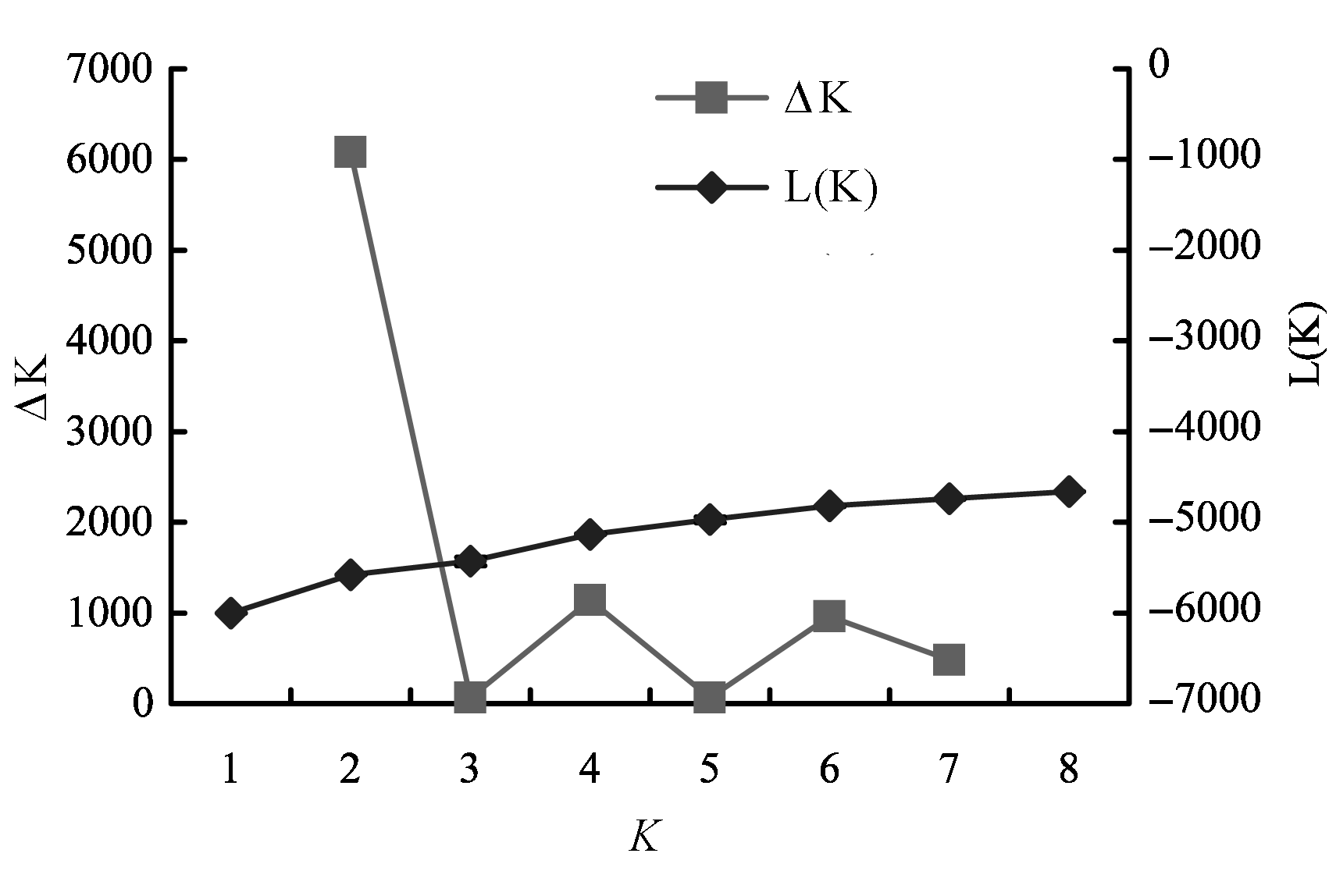
Fig. 3 Plot of mean likelihood values (averaged across 7 runs) and estimate of ΔK for each possible value of K using the data obtained from Structure software
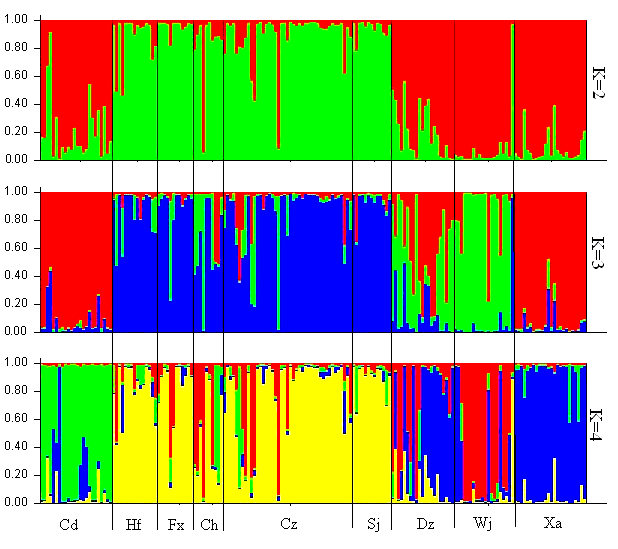
Fig. 4 Structure bar plots of Procambarus clarkii from nine local populations. The assignment results show K = 2, 3 and 4, respectively. Each vertical bar represents one individual, and sampling location codes (see Table 1) are indicated along the X-axis. Y-coordinate denotes the proportion of ancestry components in an individual in relation to other population.
| Ch | Czh | Fx | Sj | Dz | Wj | Hf | Xa | Cd | |
|---|---|---|---|---|---|---|---|---|---|
| Ch | 4.00 | 1.56 | 4.08 | 2.59 | 1.63 | 2.86 | 1.52 | 3.13 | |
| Czh | 0.06 | 1.67 | 9.49 | 2.52 | 1.62 | 5.75 | 1.38 | 1.92 | |
| Fx | 0.14 | 0.13 | 2.89 | 1.23 | 2.00 | 1.71 | 0.86 | 1.27 | |
| Sj | 0.06 | 0.03 | 0.08 | 2.72 | 2.58 | 3.97 | 1.27 | 2.03 | |
| Dz | 0.09 | 0.09 | 0.17 | 0.08 | 2.66 | 1.54 | 2.72 | 2.26 | |
| Wj | 0.13 | 0.13 | 0.11 | 0.09 | 0.09 | 1.80 | 1.92 | 2.41 | |
| Hf | 0.08 | 0.04 | 0.13 | 0.06 | 0.14 | 0.12 | 1.11 | 1.57 | |
| Xa | 0.14 | 0.15 | 0.23 | 0.16 | 0.08 | 0.12 | 0.18 | 1.69 | |
| Cd | 0.07 | 0.12 | 0.16 | 0.11 | 0.10 | 0.09 | 0.14 | 0.13 |
Table 3 Gene flow (above diagonal) and genetic diversity (Fst) (below diagonal) in nine Procambarus clarkii populations. Population codes see Table 1.
| Ch | Czh | Fx | Sj | Dz | Wj | Hf | Xa | Cd | |
|---|---|---|---|---|---|---|---|---|---|
| Ch | 4.00 | 1.56 | 4.08 | 2.59 | 1.63 | 2.86 | 1.52 | 3.13 | |
| Czh | 0.06 | 1.67 | 9.49 | 2.52 | 1.62 | 5.75 | 1.38 | 1.92 | |
| Fx | 0.14 | 0.13 | 2.89 | 1.23 | 2.00 | 1.71 | 0.86 | 1.27 | |
| Sj | 0.06 | 0.03 | 0.08 | 2.72 | 2.58 | 3.97 | 1.27 | 2.03 | |
| Dz | 0.09 | 0.09 | 0.17 | 0.08 | 2.66 | 1.54 | 2.72 | 2.26 | |
| Wj | 0.13 | 0.13 | 0.11 | 0.09 | 0.09 | 1.80 | 1.92 | 2.41 | |
| Hf | 0.08 | 0.04 | 0.13 | 0.06 | 0.14 | 0.12 | 1.11 | 1.57 | |
| Xa | 0.14 | 0.15 | 0.23 | 0.16 | 0.08 | 0.12 | 0.18 | 1.69 | |
| Cd | 0.07 | 0.12 | 0.16 | 0.11 | 0.10 | 0.09 | 0.14 | 0.13 |
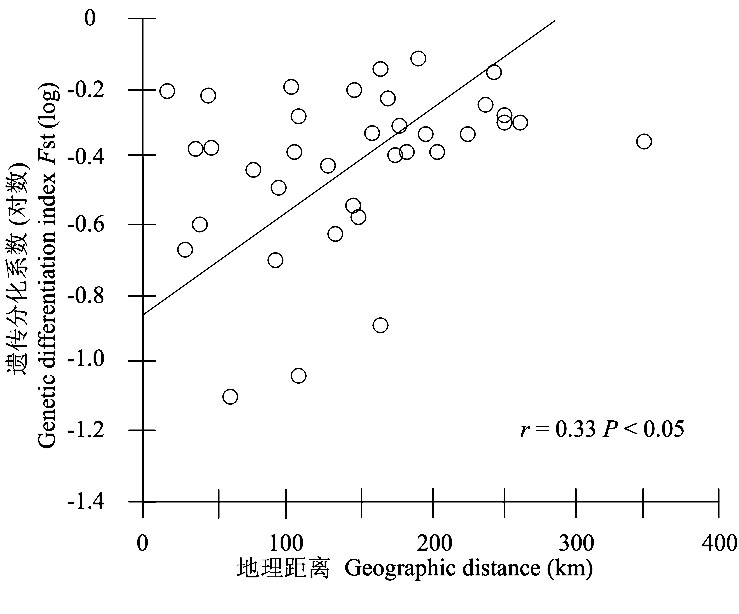
Fig. 5 Relationships between geographical distance and population genetic differentiation index (log) of nine sample sites using IBDWS. A Mantel Test indicated a significant correlation (r= 0.33, P< 0.05)
| [1] | Barbaresi S, Fani R, Gherardi F, Mengoni A, Souty-Grosset C (2003) Genetic variability of European populations of an invasive American crayfish: preliminary results. Biological Invasions, 5,269-274. |
| [2] | Barbaresi S, Gherardi F, Mengoni A, Souty-Grosset C (2007) Genetics and invasion biology in fresh waters:a pilot study of Procambarus clarkii in Europe. In: Biological Invaders in Inland Waters: Profile, Distribution and Threats (ed. Gherardi F), pp.381-400. Springer, Dordrecht. |
| [3] |
Belfiore NM, May B (2000) Variable microsatellite loci in red swamp crayfish, Procambarus clarkii, and their characterization in other crayfish taxa. Molecular Ecology, 9,2231-2234.
URL PMID |
| [4] | Berg DJ, Garton DW, Macisaac HJ, Panov VE, Telesh IV (2002) Changes in genetic structure of North American Bythotrephes populations following invasion from Lake Ladoga, Russia. Freshwater Biology, 47,275-282. |
| [5] |
Castric V, Bernatchez L, Belkhir K, Bonhomme F (2002) Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchill (Pices, Salmonidae): a test of alternative hypotheses. Heredity, 89,27-35.
URL PMID |
| [6] | Chen YF (陈毅峰), Yan YZ (严云志) (2005) Evolutionary biology of invasions. Acta Hydrobiologica Sinica (水生生物学报), 29,220-224. (in Chinese) |
| [7] | Chu D (褚栋), Zhang YJ (张友军), Wan FH (万方浩) (2007) Application of molecular marker techniques in invasion ecology. Chinese Journal of Applied Ecology (应用生态学报), 18,1383-1387. (in Chinese with English abstract) |
| [8] |
DeAssis JB, DeLaat DM, Peixoto MGCD, Bergmann JAG, Fonseca CG, Carvalho MRS (2009) Genetic diversity and population structure in Brazilian Mangalarga Marchador horses. Genetics and Molecular Research, 8,1519-1524.
DOI URL PMID |
| [9] |
Facon B, Pointier JP, Glaubrecht M, Poux C, Jarne P, David P (2003) A molecular phylogeography approach to biological invasions of the New World by parthenogenetic Thiarid snails. Molecular Ecology, 12,3027-3039.
URL PMID |
| [10] |
Friar EA, Ladoux TA, Roalson EH, Robichaux RH (2000) Microsatellite analysis of a population crash and bottleneck in the Mauna Kea silverword, Argyroxiphium sandwicense ssp. sandwicense (Asteraceae), and its implications for reintroduction. Molecular Ecology, 9,2027-2034.
DOI URL PMID |
| [11] | Gaffney PM, Scott TM, Koehn RK, Diehl WJ (1990) Interrelationships of heterozygosity, growth rate heterozygote deficiencies in the coot clam, Mutinia lateralis. Genetics and Molecular Biology, 124,687-699. |
| [12] | Geiger W, Alcorlo P, Baltanas A, Montes C (2005) Impact of an introduced Crustacean on the trophic webs of Mediterranean wetlands. Biological Invasions, 7,49-73. |
| [13] | Gherardi F (2006) Crayfish invading Europe: the case study of Procambarus clarkii. Marine and Freshwater Behaviour and Physiology, 39,175-191. |
| [14] | Gherardi F, Silvia B (2000) Invasive crayfish: activity patterns of Procambarus clarkii in the rice fields of the Lower Guadalquivir (Spain). Archiv für Hydrobiologie, 150,153-168. (in Spanish with English abstract). |
| [15] | Gregorius HR (1980) The probability of losing an allele when diploid genotype are sampled. Biometrics, 36,632-652. |
| [16] | Guo XM (郭晓鸣), Zhu SQ (朱松泉) (1997) A preliminary study on the larval development of the crayfish Procambarus clarkii. Acta Zoologica Sinica (动物学报), 43,372-381. (in Chinese with English abstract) |
| [17] | Hassan M, Bonhomme F (2005) No reduction in neutral variability of mitochondrial and nuclear genes for a Lessepsian migrant, Upeneus moluccensis. Journal of Fish Biology, 66,865-870. |
| [18] | Hedrick PW (1984) Population Biology: The Evolution and Ecology of Populations. Jones and Bartlett Publishers, Boston. |
| [19] |
Herborg LM, Weetman D, Oosterhout C, Hänfling B (2007) Genetic population structure and contemporary dispersal patterns of a recent European invader, the Chinese mitten crab, Eriocheir sinensis. Molecular Ecology, 16,231-242.
URL PMID |
| [20] | Hernández L, Maeda-Martínez AM, Ruiz-Campos G, Rodríguez-Almaraz G, Alonzo-Rojo F, Sainz JC (2007) Geographic expansion of the invasive red crayfish Procambarus clarkii (Girard, 1852) (Crustacea: Decapoda) in Mexico. Biological Invasions, 10,977-984. |
| [21] | Huner (1988) Procambarus in North America and elsewhere. In: Freshwater Crayfish: Biology, Management and Exploitation (eds Holdich DM, Lowery RS), pp.239-261. Timber Press, London. |
| [22] |
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genetics, 6,13.
URL PMID |
| [23] |
Kerby JL, Riley SPD, Kats LB, Wilson P (2005) Barriers and flow as limiting factors in the spread of an invasive crayfish (Procambarus clarkii) in southern California streams. Biological Conservation, 126,402-409.
DOI URL |
| [24] |
Kitamoto N, Honjo M, Ueno S, Takenaka A, Tsumura Y, Washitani I, Ohsawa R (2005) Spatial genetic structure among and within populations of Primula sieboldii growing beside separate streams. Molecular Ecology, 14,149-157.
DOI URL PMID |
| [25] |
Kolbe JJ, Glor RE, Schettino LR, Lara AC, Larson A, Losos JB (2004) Genetic variation increases during the biological invasion of a Cuban lizard. Nature, 431,177-181.
URL PMID |
| [26] |
Kolbe JJ, Larson A, Losos JB, Queiroz K (2008) Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biology Letters, 4,434-437.
URL PMID |
| [27] |
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, 9,299-306.
URL PMID |
| [28] | Lee CE (2002) Evolutionary genetics of invasive species. Trends in Ecology and Evolution, 17,386-391. |
| [29] | Li CC (1995) Population Genetics. The University of Chicago Press, Chicago and London. |
| [30] | Li LP (李浪平), Lü JL (吕建林), Gong SY (龚世园), Shu XY (舒新亚), Gong LJ (龚珞军) (2006) Studies on the morphology of Procambarus clarkii. Reservoir Fisheries (水利渔业), 26(3),40-42. (in Chinese) |
| [31] | Li SC (李顺才), Xu XY (徐兴友), Du LQ (杜利强), Yin XL (尹秀玲), Meng XD (孟宪东), Xie JY (解敬瑶) (2005) Investigation on and analysis of alien invasions in Chinese farming industry. Chinese Agricultural Science Bulletin (中国农学通报), 21(6),156-159. (in Chinese with English abstract) |
| [32] | Li ZY (李振宇), Xie Y (解焱) (2002) Invasive Alien Species in China (中国外来入侵种). China Forestry Publishing House, Beijing. (in Chinese) |
| [33] |
Manel S, Schwartz MK, Gordon L, Pierre T (2003) Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology and Evolution, 18,189-197.
DOI URL |
| [34] | Manning AD, Lindenmayer DB, Nix HA (2004) Continua and umwelt: novel perspectives on viewing landscapes. Oikos, 104,621-628. |
| [35] | Maria JC, Rebelo R (2007) Colonization of freshwater habitats by an introduced crayfish, Procambarus clarkii, in Southwest Iberian Peninsula. Hydrobiologia, 575,191-201. |
| [36] |
Morrison LJ, Tweedie A, Black A, Pinchbeck GL, Christley RM, Schoenefeld A, Hertz-Fowler C, MacLeod A, Turner CM, Tait A (2009) Discovery of mating in the major African livestock pathogen Trypanosoma congolense. PLoS ONE, 4,e5564.
URL PMID |
| [37] | Park SDE (2001) Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. PhD dissertation of University of Dublin, Dublin. |
| [38] | Parker IM, Simberloff D, Lonsdale WM, Goodell K, Wonham M, Kareiva PM, Williamson MH, Holle BV, Moyle PB, Byers JE, Goldwasser L (1999) Impact: toward a framework for understanding the ecological effects of invaders. Biological Invasions, 1,3-19. |
| [39] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155,945-959.
URL PMID |
| [40] | Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86,248-249. |
| [41] |
Rousset F (2008) GENEPOP' 007: a complete re-implemen- tation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources, 8,103-106.
DOI URL PMID |
| [42] |
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4,406-425.
URL PMID |
| [43] | Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O'Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Annual Review of Ecology and Systematics, 32,305-332. |
| [44] | Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York. |
| [45] |
Serrano M, Calvo JH, Martínez M, Marcos-Carcavilla A, Cuevas J, González C, Jurado JJ, de Tejada PD (2009) Microsatellite based genetic diversity and population structure of the endangered Spanish Guadarrama goat breed. BMC Genetics, 10,61.
URL PMID |
| [46] | Shi GR (史刚荣), Ma CC (马成仓) (2006) Biological characteristics of alien plants successful invasion. Chinese Journal of Applied Ecology (应用生态学报), 17,727-732. (in Chinese with English abstract) |
| [47] |
Simberloff D (2005) The politics of assessing risk for biological invasions: the USA as a case study. Trends in Ecology and Evolution, 20,216-222.
URL PMID |
| [48] | Sjogren P, Wyoni PI (1994) Conservation genetics and detection of rare alleles in finite populations. Conservation Biology, 8,267-270. |
| [49] |
Sork VL, Nason J, Campbell DR, Fernandez JF (1999) Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology and Evolution, 14,219-224.
URL PMID |
| [50] | Stepien CA, Taylor CD, Dabrowska KA (2002) Genetic variability and phylogeographical patterns of a nonindigenous species invasion: a comparison of exotic vs. native zebra and quagga mussel populations. Journal of Evolutionary Biology, 15,314-328. |
| [51] | Suarez AV, Holway DA, Case TJ (2001) Patterns of spread in biological invasions dominated by long-distance jump dispersal: insights from Argentine ants. Proceedings of the National Academy of Sciences, USA, 98,1095-1100. |
| [52] | Tiunov AV, Cindy H, Andrew H, Tamara VP (2006) Invasion patterns of Lumbricidae into the previously earthworm-free areas of northeastern Europe and the western Great Lakes region of North America. Biological Invasions, 8,1223-1234. |
| [53] |
Tsutsui ND, Suarez AV, Holway DA, Case TJ (2001) Relationships among native and introduced populations of the Argentine ant, Linepithema humile and the source of introduced populations. Molecular Ecology, 10,2151-2161.
URL PMID |
| [54] |
Valles-Jimenez R, Cruz P, Perez-Enriquez R (2005) Population genetic structure of pacific white shrimp (Litopenaeus vannamei) from Mexico to Panama microsatellite DNA variation. Marine Biotechnology, 6,475-484.
URL PMID |
| [55] |
Villanelli F, Gherardi F (1998) Breeding in the crayfish, Austropotamobius pallipes: mating patterns, mate choice and intermale competition. Freshwater Biology, 40,305-315.
DOI URL |
| [56] | Wang CZ (王长忠), Li Z (李忠), Liang HW (梁宏伟), Hu GF (呼光富), Wu QC (吴勤超), Zou GW (邹桂伟), Luo XZ (罗相忠) (2009) Genetic diversity in four Procambarus clarkii populations in the lower reaches of the Yangtze River. Biodiversity Science (生物多样性), 17,518-523. (in Chinese with English abstract) |
| [57] | Wu ZJ (武正军), Cai FJ (蔡凤金), Jia YF (贾运锋), Lu JX (鲁建鑫), Jiang YF (蒋勇福), Huang CM (黄乘明) (2008) Predation impact of Procambarus clarkii on Rana limnocharis tadpoles in Guilin area. Biodiversity Science (生物多样性), 16,150-155. (in Chinese with English abstract) |
| [58] | Xie Y (解焱), Wang S (汪松) (2001) Review on invasive species in China. In: Protecting the Biodiversity of China (II) (保护我国的生物多样性) (eds Schei P, Xie Y (解焱), Wang S (汪松)), pp.91-96. China Environmental Science Press, Beijing. (in Chinese) |
| [59] | Xu RM (徐汝梅), Ye WH (叶万辉) (2003) Biological Invasion: Theory and Practice (生物入侵: 理论与实践). Science Press, Beijing. (in Chinese) |
| [60] | Yan LN (闫路娜), Zhang DX (张德兴) (2004) Effects of sample size on various genetic diversity measures in population genetic study with microsatelite DNA markers. Acta Zoologica Sinica (动物学报), 50,279-290. (in Chinese with English abstract) |
| [61] |
Yasuda N, Nagai S, Hamaguchi M, Okaji K, Gérard K, Nadaoka K (2009) Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Molecular Ecology, 18,1574-1590.
DOI URL PMID |
| [62] | Yue GH, Li JL, Bai ZY, Wang CM, Feng F (2010) Genetic diversity and population structure of the invasive alien red swamp crayfish. Biological Invasions, 12,2697-2706. |
| [63] |
Zalewski A, Piertney SB, Zalewska H, Lambin X (2009) Landscape barriers reduce gene flow in an invasive carnivore: geographical and local genetic structure of American mink in Scotland. Molecular Ecology, 18,1601-1615.
DOI URL PMID |
| [64] |
Zhu ZY, Yue GH (2008) Eleven polymorphic microsatellites isolated from red swamp crayfish, Procambarus clarkii. Molecular Ecology Resources, 8,796-798.
URL PMID |
| [1] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [2] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [3] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [4] | Congcong Du, Xueyu Feng, Zhilin Chen. The reducing of climate niche differences in the bridgehead effect promotes the invasion of Solenopsis invicta [J]. Biodiv Sci, 2024, 32(11): 24276-. |
| [5] | Xuejiao Yuan, Yuanyuan Zhang, Yanliang Zhang, Luyi Hu, Weiguo Sang, Zheng Yang, Qi Chen. Investigating the prediction ability of the species distribution model fitted with the historical distribution records of Chromolaena odorata [J]. Biodiv Sci, 2024, 32(11): 24288-. |
| [6] | Lixia Han, Yongjian Wang, Xuan Liu. Comparisons between non-native species invasion and native species range expansion [J]. Biodiv Sci, 2024, 32(1): 23396-. |
| [7] | Jiajia Pu, Pingjun Yang, Yang Dai, Kexin Tao, Lei Gao, Yuzhou Du, Jun Cao, Xiaoping Yu, Qianqian Yang. Species identification and population genetic structure of non-native apple snails (Ampullariidea: Pomacea) in the lower reaches of the Yangtze River [J]. Biodiv Sci, 2023, 31(3): 22346-. |
| [8] | Bo Wei, Linshan Liu, Changjun Gu, Haibin Yu, Yili Zhang, Binghua Zhang, Bohao Cui, Dianqing Gong, Yanli Tu. The climate niche is stable and the distribution area of Ageratina adenophora is predicted to expand in China [J]. Biodiv Sci, 2022, 30(8): 21443-. |
| [9] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [10] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [11] | Yanjie Liu, Wei Huang, Qiang Yang, Yu-Long Zheng, Shao-Peng Li, Hao Wu, Ruiting Ju, Yan Sun, Jianqing Ding. Research advances of plant invasion ecology over the past 10 years [J]. Biodiv Sci, 2022, 30(10): 22438-. |
| [12] | Jing Yan, Xiaoling Yan, Huiru Li, Cheng Du, Jinshuang Ma. Composition, time of introduction and spatial-temporal distribution of naturalized plants in East China [J]. Biodiv Sci, 2021, 29(4): 428-438. |
| [13] | Weiming He. Biological invasions: Are their impacts precisely knowable or not? [J]. Biodiv Sci, 2020, 28(2): 253-255. |
| [14] | Jiazhen Zhang, Chunlei Gao, Yan Li, Ping Sun, Zongling Wang. Species composition of dinoflagellates cysts in ballast tank sediments of foreign ships berthed in Jiangyin Port [J]. Biodiv Sci, 2020, 28(2): 144-154. |
| [15] | Wandong Yin, Mingke Wu, Baoliang Tian, Hongwei Yu, Qiyun Wang, Jianqing Ding. Effects of bio-invasion on the Yellow River basin ecosystem and its countermeasures [J]. Biodiv Sci, 2020, 28(12): 1533-1545. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()