



Biodiv Sci ›› 2009, Vol. 17 ›› Issue (5): 490-498. DOI: 10.3724/SP.J.1003.2009.09101 cstr: 32101.14.SP.J.1003.2009.09101
• Editorial • Previous Articles Next Articles
Xinxin Sun, Huirong Liu, Fuying Feng( ), Jianyu Meng, Heng Li, Malina
), Jianyu Meng, Heng Li, Malina
Received:2009-04-16
Accepted:2009-06-24
Online:2009-09-20
Published:2009-09-20
Contact:
Fuying Feng
Xinxin Sun, Huirong Liu, Fuying Feng, Jianyu Meng, Heng Li, Malina. Diversity and phylogenetic analysis of planktonic bacteria in eutrophic zone of Lake Wuliangsuhai[J]. Biodiv Sci, 2009, 17(5): 490-498.
| 铵态氮 NH4+-N (mg/L) | 硝态氮 NO3--N (mg/L) | 总氮 Total N (mg/L) | 总磷 Total P (mg/L) | 硫化物 S (mg/L) | Na+ Na+ (mg/L) | 化学需氧量 CODMn (mg/L) | 溶解有机碳 DOC (mg/L) | 叶绿素a Chl a (mg/L) | 温度T Temperature (℃) | pH | 细菌数量 Bacterial number (cells/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.79 | 0.158 | 4.87 | 0.247 | 0.364 | 463 | 69.21 | 85.95 | 13.8 | 24.2 | 8.72 | 6.58×107 |
Table 1 Water parameters and total bacterial numbers in Wuliangsuhai Lake
| 铵态氮 NH4+-N (mg/L) | 硝态氮 NO3--N (mg/L) | 总氮 Total N (mg/L) | 总磷 Total P (mg/L) | 硫化物 S (mg/L) | Na+ Na+ (mg/L) | 化学需氧量 CODMn (mg/L) | 溶解有机碳 DOC (mg/L) | 叶绿素a Chl a (mg/L) | 温度T Temperature (℃) | pH | 细菌数量 Bacterial number (cells/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.79 | 0.158 | 4.87 | 0.247 | 0.364 | 463 | 69.21 | 85.95 | 13.8 | 24.2 | 8.72 | 6.58×107 |
| OTU代表 Representatives of OTU | 克隆数 Clone number | RDP属 RDP genus | |
|---|---|---|---|
| 属 Genus | 置信度 Confidence(%) | ||
| H0110 | 5 | Rhodobacter | 100 |
| H0015 | 4 | NC | 12 |
| H0080 | 5 | Hydrogenophaga | 100 |
| H0022 | 1 | Hydrogenophaga | 100 |
| H0061 | 1 | Curvibacter | 88 |
| H0108 | 5 | NC | 77 |
| H0035 | 9 | NC | 29 |
| H0051 | 9 | Curvibacter | 98 |
| H0074 | 6 | Methylophilus | 90 |
| H0068 | 3 | Aeromonas | 100 |
| H0030 | 1 | Shigella | 99 |
| H0041 | 10 | NC | 23 |
| H0002 | 3 | NC | 51 |
| H0017 | 2 | Fluviicola | 100 |
| H0093 | 1 | Flavobacterium | 100 |
| H0097 | 1 | NC | 67 |
| H0052 | 4 | Terrimonas | 96 |
| H0091 | 2 | NC | 33 |
| H0050 | 4 | NC | 76 |
| H0064 | 2 | NC | 31 |
| H0054 | 2 | Xi | 100 |
| H0057 | 4 | Exiguobacterium | 100 |
| H0011 | 3 | NC | 58 |
Table 2 Digestion patterns and proportion of 16S rDNA by HaeIII and classification
| OTU代表 Representatives of OTU | 克隆数 Clone number | RDP属 RDP genus | |
|---|---|---|---|
| 属 Genus | 置信度 Confidence(%) | ||
| H0110 | 5 | Rhodobacter | 100 |
| H0015 | 4 | NC | 12 |
| H0080 | 5 | Hydrogenophaga | 100 |
| H0022 | 1 | Hydrogenophaga | 100 |
| H0061 | 1 | Curvibacter | 88 |
| H0108 | 5 | NC | 77 |
| H0035 | 9 | NC | 29 |
| H0051 | 9 | Curvibacter | 98 |
| H0074 | 6 | Methylophilus | 90 |
| H0068 | 3 | Aeromonas | 100 |
| H0030 | 1 | Shigella | 99 |
| H0041 | 10 | NC | 23 |
| H0002 | 3 | NC | 51 |
| H0017 | 2 | Fluviicola | 100 |
| H0093 | 1 | Flavobacterium | 100 |
| H0097 | 1 | NC | 67 |
| H0052 | 4 | Terrimonas | 96 |
| H0091 | 2 | NC | 33 |
| H0050 | 4 | NC | 76 |
| H0064 | 2 | NC | 31 |
| H0054 | 2 | Xi | 100 |
| H0057 | 4 | Exiguobacterium | 100 |
| H0011 | 3 | NC | 58 |
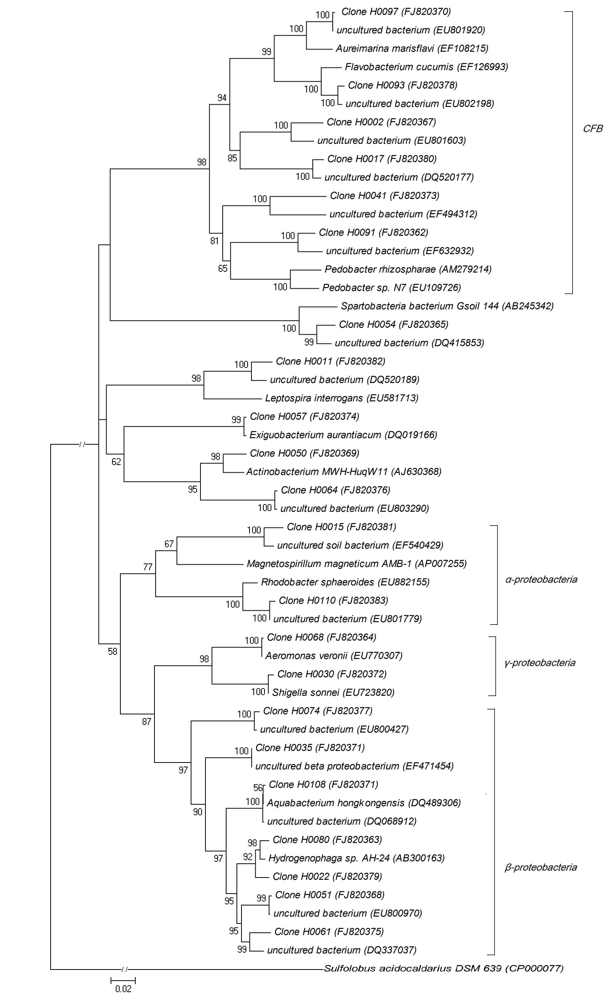
Fig. 1 Phylogenetic tree of bacterial 16S rDNA gene sequences in Wuliangsuhai Lake water. Data in parentheses are the GenBank accession numbers. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1,000 resampled datasets. Sulfolobus acidocaldarius DSM 639 serves as outgroup.
| [1] |
Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF (2005) PCR-induced sequence artifacts and bias: insights from comparision of two 16S rRNA clone libraries constructed from the same sample. Applied and Environmental Microbiology, 71,8966-8969.
DOI URL PMID |
| [2] | Amann RI, Ludwing W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiologyical Reviews, 59,143-169. |
| [3] | Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2002) Short Protocols in Molecular Biology. Wiley Press, New York. |
| [4] | Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, MacGarrell DM, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research, 37,D141-D145. |
| [5] |
Cottrell MT, Waidner LA, Yu LY, Kirchman DL (2005) Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environmental Microbiology, 7,1883-1895.
DOI URL PMID |
| [6] |
Demergasso C, Casamayor EO, Chong G, Galleguillos P, Escudero L, Pedros-Alio C (2004) Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiology Ecology, 48,57-69.
DOI URL PMID |
| [7] |
Dimitriu PA, Pinkart HC, Peyton BM, Mormile M (2008) Spatial and temporal patterns in the microbial diversity of a meromictic soda lake in Washington State. Applied and Environmental Microbiology, 74,4877-4888.
DOI URL PMID |
| [8] | Duan XN (段晓男), Wang XK (王效科), Ouyang ZY (欧阳志云) (2005) Evaluation of wetland ecosystems services in Wuliangsuhai. Resources Science (资源科学), 27(2),110-115. (in Chinese with English abstract) |
| [9] | Felsenstein J (1989) PHYLIP–Phylogeny Inference Package (Version 3.2). Cladistics, 5,164-166. |
| [10] | Feng S (冯胜), Qin BQ (秦伯强), Gao G (高光) (2007) Response of bacterial communities to eutrophic water in Lake Taihu. Acta Scientiae Circumstantiae (环境科学学报), 27,1823-1829. (in Chinese with English abstract) |
| [11] |
Frostegard A, Courtois S, Ramisse V, Clerc S, Bernillon D, Le Gall F, Jeannin P, Nesme X, Simonet P (1999) Quantification of bias related to the extraction of DNA directly from soils. Applied and Environmental Microbiology, 65,5409-5420.
DOI URL PMID |
| [12] |
Giovannoni SJ, Sting U (2005) Molecular diversity and ecology of microbial plankton. Nature, 437,343-348.
DOI URL PMID |
| [13] |
Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amannl R (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Applied and Environmental Microbiology, 66,5053-5065.
URL PMID |
| [14] | Grant S, Sorokin DY, Grant WD, Jones BE, Heaphy S (2004) A phylogenetic analysis of Wadi el Natrun soda lake cellulose enrichment cultures. Extremephiles, 8,421-429. |
| [15] |
Hill TCJ, Walsh KA, Harris JA, Moffett BF (2003) Using ecological diversity measures with bacterial communities. FEMS Microbiology Ecology, 43,1-11.
DOI URL PMID |
| [16] | Jenkins O, Byrom D, Jones D (1987) Methylophilus: a new genus of methanol-utilizing bacteria. International Journal of Systematic Bacteriology, 37,446-448. |
| [17] |
Kämpfer P, Schulze R, Jackel U, Malik KA, Amann R, Spring S (2005) Hydrogenophaga defluvii sp. nov. and Hydrogenophaga atypica sp. nov., isolated from activated sludge. International Journal of Systematic and Evolutionary Microbiology, 55,341-344.
DOI URL PMID |
| [18] |
Kumar S, Dudley J, Nei M, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, 9,299-306.
DOI URL PMID |
| [19] | Li CY (李畅游), Gao RZ (高瑞忠), Liu TX (刘廷玺), Ren CT (任春涛) (2005) Study on the eutrophication synthetical evaluation and the season-year change in Wuliangsuhai Lake. Journal of Water Resources and Water Engineering (水资源与水工程学报), 16(2),11-15. (in Chinese with English abstract) |
| [20] |
Lindström ES, Agterveld K-V MP, Zwart G (2005) Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Applied and Environmental Microbiology, 71,8201-8206.
DOI URL PMID |
| [21] | Liu YQ, Yao TD, Zhu LP, Jiao NZ, Liu XB, Zeng YH, Jiang HC (2009) Bacterial diversity of freshwater alpine lake Puma Yumco on the Tibetan Plateau. Geomicrobiology Journal, 26,131-145. |
| [22] |
Lovejoy C, Massana R, Pedrós-Alió C (2006) Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Applied and Environmental Microbiology, 72,3085-3095.
DOI URL PMID |
| [23] |
Ma YH, Zhang WZ, Xue YF, Zhou PJ, Ventosa A, Grant WD (2004) Bacterial diversity of the Inner Mongolian Baer soda lake as revealed by 16S rRNA gene sequence analyses. Extremophiles, 8,45-51.
DOI URL PMID |
| [24] |
MacCann KS (2000) The diversity-stability debate. Nature, 405,228-233.
URL PMID |
| [25] |
Moyer CL, Tiedje JM, Dobbs FC (1996) A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected terameric restriction enzymes for studies of microbial diversity in nature. Applied and Environmental Microbiology, 62,2501-2507.
DOI URL PMID |
| [26] |
Oda Y, Wanders W, Huisman LA, Meijer WG, Gottschal JC, Forney LJ (2002) Genotypic and phenotypic diversity within species of purple nonsulfur bacteria isolated from aquatic sediments. Applied and Environmental Microbiology, 68,3467-3477.
DOI URL PMID |
| [27] |
Percent SF, Frischer ME, Vescio PA, Duffy VE, Milano V, McLellan M, Stevens BM, Boylen CW, Nierzwicki-Bauer SA (2008) Bacterial community structure of acid-impacted lakes: what controls diversity? Applied and Environmental Microbiology, 74,1856-1868.
DOI URL PMID |
| [28] | Qin BQ, Yang LY, Chen FZ, Zhu GW, Zhang L, Chen YY (2006) Mechanism and control of lake eutrophication. Chinese Science Bulletin, 51,2401-2402. |
| [29] | Qu JH (屈建航), Yuan HL (袁红莉), Huang HC (黄怀曾), Wang ET (汪恩涛) (2005) Vertical distribution of bacterial community in sediment of Guantin Reservoir. Science in China Series D Earth Science(Suppl. I), 35,233-240. (in Chinese) |
| [30] |
Rodrigues DF, Goris J, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM (2006) Characterization of Exiguobacterium isolates from the Siberian permafrost. Description of Exiguobacterium sibiricum sp. nov. Extremophiles, 10,285-294.
DOI URL PMID |
| [31] |
Sekiguchi H, Watanabe M, Nakahara T, Xu BH, Uchiyama H (2002) Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Applied and Environmental Microbiology, 68,5142-5150.
DOI URL PMID |
| [32] |
Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JBH (2008) It’s all relative: ranking the diversity of aquatic bacterial communities. Environmental Microbiology, 10,2200-2210.
DOI URL PMID |
| [33] | Shi XH (史小红) (2007) Modeling Analysis of Eutrophic Elements and Their Speciation for Wuliangsuhai Lake in Inner Mongolia (乌梁素海营养元素及其存在形态的数值模拟分析). PhD dissertation, Inner Mongolia Agriculture University, Huhhot. (in Chinese with English summary). |
| [34] |
Sipos R, Székely AJ, Palatinszky M, Révész S, Márialigeti K, Nikolausz M (2007) Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targeting bacterial community analysis. FEMS Microbiology Ecology, 60,341-350.
DOI URL PMID |
| [35] |
Tamaki H, Sekiguchi Y, Hanada S, Nakamura K, Nomura N, Matsumura M, Kamagata Y (2005) Comparative analysis of bacteria diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Applied and Environmental Microbiology, 71,2162-2169.
DOI URL PMID |
| [36] |
Tiago I, Chung AP, Verissimo A (2004) Bacterial diversity in a nonsaline alkaline environment: heterotrophic aerobic populations. Applied and Environmental Microbiology, 70,7378-7387.
DOI URL PMID |
| [37] |
Vieira RP, Gonzalez AM, Cardoso AM, Oliverira DN, Albano RM, Clementino MM, Martins OB, Paranhos R (2008) Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environmental Microbiology, 10,189-199.
DOI URL PMID |
| [38] |
von Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiology Reviews, 21,213-229.
DOI URL PMID |
| [39] | Wang LM (王丽敏), Shang SY (尚士友), Wu LB (吴利斌), Yue HJ (岳海军) (2004) Study on the nitrogen transformation in Wuliangsuhai, a grass-type lake. Environmental Science Trends (环境科学动态), (1),16-18. (in Chinese) |
| [40] | Wang ZW, Liu YH, Dai X, Wang BJ, Jiang CY, Liu SJ (2006) Flavobacterium saliperosum sp. nov., isolated from freshwater lake sediment. International Journal of Systematic Bacteriology, 56,439-442. |
| [41] |
Wu X, Xi WY, Ye WJ, Yang H (2007) Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences. FEMS Microbiology Ecology, 61,85-96.
URL PMID |
| [42] | Yachi S, Loreau M (1999) Biodiversity and ecosystem functioning in a fluctuating environment: the insurance hypothesis. Proceedings of the National Academy Sciences USA, 96,1463-1468. |
| [43] |
Yoon KS, Tsukada N, Sakai Y, Ishii M, Igarashi Y, Nishihara H (2008) Isolation and characterization of a new facultatively autotrophic hydrogen-oxidizing beta-proteobacterium, Hydrogenophaga sp. AH-24. FEMS Microbiology Letters, 278,94-100.
DOI URL PMID |
| [44] | Zhang WZ (张伟周), Mao WY (毛文扬), Xue YF (薛燕芬), Ma YH (马延和), Zhou PJ (周培瑾) (2001) The diversity of alkaliphiles from Hailaer Soda Lake, Inner Mongolia. Biodiversity Science (生物多样性), 9,44-50. (in Chinese with English abstract) |
| [45] | Zwart G, Crump BC, Agterveld M, Hagen F, Han SK (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquatic Microbial Ecology, 28,141-155. |
| [1] | Zhiyu Liu, Xin Ji, Guohui Sui, Ding Yang, Xuankun Li. Invertebrate diversity in buffalo grass and weedy lawns at Beijing Capital International Airport [J]. Biodiv Sci, 2025, 33(4): 24456-. |
| [2] | Li Hualiang, Zhang Mingjun, Zhang Xibin, Tan Rong, Li Shichuan, Feng Erhui, Lin Xueyun, Chen Min, Yan enbo, Zeng Zhigao. Composition and influencing factors of the amphibian community in Hainan Dongzhaigang National Nature Reserve [J]. Biodiv Sci, 2025, 33(2): 24350-. |
| [3] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [4] | Xinyang Zhou, Yutao Wang, Jianping Li. Response of plant community composition to precipitation changes in typical grasslands in the Loess Plateau [J]. Biodiv Sci, 2023, 31(3): 22118-. |
| [5] | Chaoya Wang, Jintao Li, Chang Liu, Bo Wang, Baige Miao, Yanqiong Peng. Interannual stability in butterfly diversity and the larvae-plant interaction network structure at Xishuangbanna Tropical Botanical Garden [J]. Biodiv Sci, 2023, 31(12): 23305-. |
| [6] | Jinhua Liu, Feng Li, Tao Tian, Haifeng Xiao. Response of soil bacteria and nematodes to litter identity and diversity of dominant plants in a tropical rainforest [J]. Biodiv Sci, 2023, 31(11): 23276-. |
| [7] | Chao Zhang, Juan Li, Haiyun Cheng, Jiachong Duan, Zhao Pan. Patterns and environmental drivers of the butterfly diversity in the western region of Qinling Mountains [J]. Biodiv Sci, 2023, 31(1): 22272-. |
| [8] | Xiaotong Liu, Yijia Tian, Hanwen Liu, Cuiying Liang, Siwei Jiang, Wenju Liang, Xiaoke Zhang. Seasonal variation in cropland soil nematode community composition in the lower reaches of Liaohe Plain [J]. Biodiv Sci, 2022, 30(12): 22222-. |
| [9] | Qi Zhao, Jibao Jiang, Zenglu Zhang, Qing Jin, Jiali Li, Jiangping Qiu. Species composition and phylogenetic analysis of earthworms on Hainan Island [J]. Biodiv Sci, 2022, 30(12): 22224-. |
| [10] | Yu Ren, Shengli Tao, Tianyu Hu, Haitao Yang, Hongcan Guan, Yanjun Su, Kai Cheng, Mengxi Chen, Huawei Wan, Qinghua Guo. The outlook and system construction for monitoring Essential Biodiversity Variables based on remote sensing: The case of China [J]. Biodiv Sci, 2022, 30(10): 22530-. |
| [11] | Dongmei Li, Weihong Yang, Qingduo Li, Xi Han, Xiuping Song, Hong Pan, Yun Feng. High prevalence and genetic variation of Bartonella species inhabiting the bats in southwestern Yunnan [J]. Biodiv Sci, 2021, 29(9): 1245-1255. |
| [12] | Zusheng Yi, Yuanjun Huang, Hui Yi, Xinwang Zhang, Wenjun Li. Biodiversity of macrozoobenthos in the Chebaling National Nature Reserve, Guangdong Province [J]. Biodiv Sci, 2021, 29(5): 680-687. |
| [13] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| [14] | Zhiliang Yao,Handong Wen,Yun Deng,Min Cao,Luxiang Lin. Driving forces underlying the beta diversity of tree species in subtropical mid-mountain moist evergreen broad-leaved forests in Ailao Mountains [J]. Biodiv Sci, 2020, 28(4): 445-454. |
| [15] | Zhixiang Chen, Xueying Yao, R. Downie Stephen, Qizhi Wang. Assembling and analysis of Sanicula orthacantha chloroplast genome [J]. Biodiv Sci, 2019, 27(4): 366-372. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()