



Biodiv Sci ›› 2009, Vol. 17 ›› Issue (5): 476-481. DOI: 10.3724/SP.J.1003.2009.09100 cstr: 32101.14.SP.J.1003.2009.09100
• Editorial • Previous Articles Next Articles
Longqian Xiao1,2, Hua Zhu1,*( )
)
Received:2009-04-16
Accepted:2009-09-05
Online:2009-09-20
Published:2009-09-20
Contact:
Hua Zhu
Longqian Xiao, Hua Zhu. Intra-genomic polymorphism in the internal transcribed spacer (ITS) regions of Cycas revoluta: evidence of incomplete concerted evolution[J]. Biodiv Sci, 2009, 17(5): 476-481.
| 克隆编号 Clone No. | GenBank序列号 GenBank accession no. | 长度(GC含量%) Length (bp) (GC content %) | 5.8S基序5.8S motif | 最小自由能5.8S Δ G (kcal/mol) | ||
|---|---|---|---|---|---|---|
| ITS1 | ITS2 | 5.8S | ||||
| D1 | FJ907980 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D2 | FJ907972 | 677(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D3 | FJ908060 | 674(51.0) | 243(46.1) | 161(44.7) | V | -13.79 |
| D4 | FJ907973 | 677(63.8) | 246(65.9) | 161(55.9) | V | -16.39 |
| D5 | FJ907974 | 677(64.1) | 246(65.0) | 161(55.9) | V | -16.39 |
| D6 | FJ908059 | 676(49.4) | 243(49.4) | 161(43.5) | V | -8.54 |
| D7 | FJ908058 | 598(49.3) | 243(46.1) | 160(43.1) | -11.51 | |
| D8 | FJ908051 | 672(51.0) | 243(50.6) | 161(37.9) | -10.95 | |
| D9 | FJ907982 | 676(63.8) | 246(65.0) | 161(55.9) | V | -16.39 |
| D11 | FJ908056 | 679(50.5) | 242(50.0) | 161(42.9) | -9.42 | |
| D12 | FJ907981 | 677(64.0) | 246(65.0) | 161(55.3) | V | -16.90 |
| D13 | FJ907983 | 674(62.7) | 246(62.6) | 161(54.7) | V | -19.39 |
| D14 | FJ908045 | 658(50.2) | 243(50.6) | 161(43.5) | -10.37 | |
| D15 | FJ908067 | 671(48.7) | 243(50.2) | 161(44.7) | V | -13.21 |
| C1 | FJ907976 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| C2 | FJ907979 | 678 (64.0) | 246(65.0) | 161(55.9) | V | -16.39 |
| C3 | FJ907978 | 679(64.1) | 246(65.0) | 161(55.3) | V | -16.59 |
| C4 | FJ907977 | 677 (64.1) | 246(64.6) | 161(55.9) | V | -16.39 |
| C5 | FJ907975 | 681(64.2) | 230(65.6) | 161(55.9) | V | -16.39 |
Table 1 The characteristics of ITS1, 5.8S and ITS2 in Cycas revoluta
| 克隆编号 Clone No. | GenBank序列号 GenBank accession no. | 长度(GC含量%) Length (bp) (GC content %) | 5.8S基序5.8S motif | 最小自由能5.8S Δ G (kcal/mol) | ||
|---|---|---|---|---|---|---|
| ITS1 | ITS2 | 5.8S | ||||
| D1 | FJ907980 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D2 | FJ907972 | 677(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D3 | FJ908060 | 674(51.0) | 243(46.1) | 161(44.7) | V | -13.79 |
| D4 | FJ907973 | 677(63.8) | 246(65.9) | 161(55.9) | V | -16.39 |
| D5 | FJ907974 | 677(64.1) | 246(65.0) | 161(55.9) | V | -16.39 |
| D6 | FJ908059 | 676(49.4) | 243(49.4) | 161(43.5) | V | -8.54 |
| D7 | FJ908058 | 598(49.3) | 243(46.1) | 160(43.1) | -11.51 | |
| D8 | FJ908051 | 672(51.0) | 243(50.6) | 161(37.9) | -10.95 | |
| D9 | FJ907982 | 676(63.8) | 246(65.0) | 161(55.9) | V | -16.39 |
| D11 | FJ908056 | 679(50.5) | 242(50.0) | 161(42.9) | -9.42 | |
| D12 | FJ907981 | 677(64.0) | 246(65.0) | 161(55.3) | V | -16.90 |
| D13 | FJ907983 | 674(62.7) | 246(62.6) | 161(54.7) | V | -19.39 |
| D14 | FJ908045 | 658(50.2) | 243(50.6) | 161(43.5) | -10.37 | |
| D15 | FJ908067 | 671(48.7) | 243(50.2) | 161(44.7) | V | -13.21 |
| C1 | FJ907976 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| C2 | FJ907979 | 678 (64.0) | 246(65.0) | 161(55.9) | V | -16.39 |
| C3 | FJ907978 | 679(64.1) | 246(65.0) | 161(55.3) | V | -16.59 |
| C4 | FJ907977 | 677 (64.1) | 246(64.6) | 161(55.9) | V | -16.39 |
| C5 | FJ907975 | 681(64.2) | 230(65.6) | 161(55.9) | V | -16.39 |
| ITS区域 ITS region | 序列类型 Sequence type | 序列数目 Sequence number | 多态位点数Polymorphic site no. | 总突变数 Total mutation | 核苷酸多态性 Nucleotide diversity (π) |
|---|---|---|---|---|---|
| ITS1 | cDNA ITS | 5 | 11 | 11 | 0.007 |
| 功能 ITS Functional ITS | 7 | 25 | 25 | 0.011 | |
| 假基因 Pseudogene | 7 | 258 | 283 | 0.191 | |
| ITS2 | cDNA ITS | 5 | 2 | 2 | 0.003 |
| 功能 ITS Functional ITS | 7 | 11 | 11 | 0.013 | |
| 假基因 Pseudogene | 7 | 120 | 142 | 0.222 | |
| 5.8S | cDNA ITS | 5 | 1 | 1 | 0.002 |
| 功能 ITS Functional ITS | 7 | 4 | 4 | 0.007 | |
| 假基因 Pseudogene | 7 | 64 | 72 | 0.174 | |
| ITS | cDNA ITS | 5 | 14 | 14 | 0.005 |
| 功能 ITS Functional ITS | 7 | 40 | 40 | 0.011 | |
| 假基因 Pseudogene | 7 | 442 | 497 | 0.196 |
Table 2 Nucleotide diversity of the ITS region in Cycas revoluta
| ITS区域 ITS region | 序列类型 Sequence type | 序列数目 Sequence number | 多态位点数Polymorphic site no. | 总突变数 Total mutation | 核苷酸多态性 Nucleotide diversity (π) |
|---|---|---|---|---|---|
| ITS1 | cDNA ITS | 5 | 11 | 11 | 0.007 |
| 功能 ITS Functional ITS | 7 | 25 | 25 | 0.011 | |
| 假基因 Pseudogene | 7 | 258 | 283 | 0.191 | |
| ITS2 | cDNA ITS | 5 | 2 | 2 | 0.003 |
| 功能 ITS Functional ITS | 7 | 11 | 11 | 0.013 | |
| 假基因 Pseudogene | 7 | 120 | 142 | 0.222 | |
| 5.8S | cDNA ITS | 5 | 1 | 1 | 0.002 |
| 功能 ITS Functional ITS | 7 | 4 | 4 | 0.007 | |
| 假基因 Pseudogene | 7 | 64 | 72 | 0.174 | |
| ITS | cDNA ITS | 5 | 14 | 14 | 0.005 |
| 功能 ITS Functional ITS | 7 | 40 | 40 | 0.011 | |
| 假基因 Pseudogene | 7 | 442 | 497 | 0.196 |
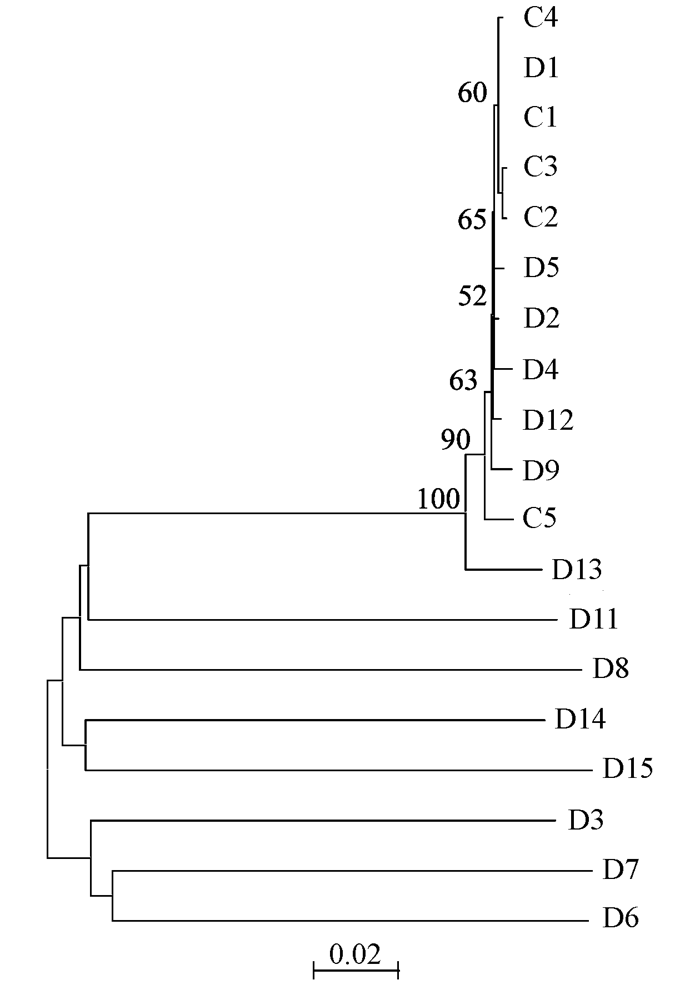
Fig. 1 Neighbor-joining tree topology constructed on Kimura two-parameter distance matrix using ITS entire region of all paralogs in C. revoluta. Numbers indicate the over 50% bootstrap values with 2,000 replicates.
| [1] |
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution, 29,417-434.
DOI URL PMID |
| [2] |
Baker WJ, Hedderson TA, Dransfield J (2000) Molecular phylogenetics of subfamily Calamoideae (Palmae) based on nrDNA ITS and cpDNA rps16 intron sequence data. Molecular Phylogenetics and Evolution, 14,195-217.
URL PMID |
| [3] | Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA―a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden, 82,247-277. |
| [4] |
Buckler ES, Holtsford TP (1996) Zea ribosomal repeat evolution and mutation patterns. Molecular Biology and Evolution, 13,623-632.
DOI URL PMID |
| [5] |
Buckler ES, Ippolito A, Holtsford TP (1997) The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics, 145,821-832.
URL PMID |
| [6] |
Campbell CS, Wright WA, Cox M, Vining TF, Major CS, Arsenault MP (2005) Nuclear ribosomal DNA internal transcribed spacer 1 (ITS1) in Picea (Pinaceae), sequence divergence and structure. Molecular Phylogenetics and Evolution, 35,165-185.
URL PMID |
| [7] |
Chaw SM, Walters TW, Chang CC, Hu SH, Chen SH (2005) A phylogeny of cycads (Cycadales) inferred from chloroplast matk gene, trnK intron, and nuclear rDNA ITS region. Molecular Phylogenetics and Evolution, 37,214-234.
DOI URL PMID |
| [8] | Doyle J (1991) DNA protocols for plants - CTAB total DNA isolation. In: Molecular Techniques in Taxonomy (eds Hewitt GM, Johnston A), pp.283-293. Springer, Berlin. |
| [9] |
Eickbush TH, Eickbush DG (2007) Finely orchestrated movements, evolution of the ribosomal RNA genes. Genetics, 175,477-485.
DOI URL PMID |
| [10] | Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41,95-98. |
| [11] |
Harpke D, Peterson A (2006) Non-concerted ITS evolution in Mammillaria (Cactaceae). Molecular Phylogenetics and Evolution, 41,579-593.
DOI URL PMID |
| [12] | Hizume M, Ishida F, Kondo K (1992) Differential staining and in situ hybridization of nucleolar oraganizers and centromeres in Cycas revoluta chromosomes. Japanese Journal of Genetics, 67,381-387. |
| [13] | Jobes DV, Thien LB (1997) A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. Plant Molecular Biology Reporter, 15,326-334. |
| [14] | Johnson LAS, Wilson KL (1990) General traits of the Cycadales. In: The Families and Genera of Vascular Plants (eds Kramer KU, Green PS), pp.363-368. Springer-Verlag, Berlin. |
| [15] |
Kan XZ, Wang SS, Ding X, Wang XQ (2007) Structural evolution of nrDNA ITS in Pinaceae and its phylogenetic implications. Molecular Phylogenetics and Evolution, 44,765-777.
DOI URL PMID |
| [16] | Karvonen P, Savolainen O (1993) Variation and inheritance of ribosomal DNA in Pinus sylvestris L. (Scots pine). Heredity, 71,614-622. |
| [17] | Kumar S, Tamura K, Nei M (1994) MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Bioinformatics, 10,189-191. |
| [18] | Liston A, Robinson WA, Oliphant JM, Alvarez-Buylla ER (1996) Length variation in the nuclear ribosomal DNA internal transcribed spacer region of non-flowering seed plants. Systematic Botany, 21,109-120. |
| [19] |
Mayol M, Rossello JA (2001) Why nuclear ribosomal DNA spacers (ITS) tell different stories in Quercus. Molecular Phylogenetics and Evolution, 19,167-176.
DOI URL PMID |
| [20] |
Muir G, Fleming CC, Schlotterer C (2001) Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Molecular Biology and Evolution, 18,112-119.
URL PMID |
| [21] |
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annual Review of Genetics, 39,121-152.
DOI URL PMID |
| [22] | Nickrent DL, Schuette KP, Starr EM (1994) A molecular phylogeny of Arceuthbium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany, 81,1149-1160. |
| [23] | Osborne R (1995) The world cycad census and a proposed revision of the threatened species status for cycad taxa. Biological Conservation, 71,1-12. |
| [24] |
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology, 265,218-225.
DOI URL PMID |
| [25] |
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics, 15,174-175.
URL PMID |
| [26] | Sang T, Crawford J, Stuessy TF (1995) Documentation of reticulate evolution in peonies ( Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA, implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences, USA, 92,6813-6817. |
| [27] | Shimabuku K (1997) Check List Vascular Flora of the Ryukyu Islands, pp.1-855. Kyushu University Press, Fukuoka. (in Japanese). |
| [28] | Stevenson DW (1990) Morphology and systematics of the Cycadales. Memoirs of the New York Botanical Garden, 57,8-55. |
| [29] |
Suh Y, Thien LB, Zimmer EA (1992) Nucleotide sequences of the internal transcribed spacers and 5.8S rRNA gene in Canella winterana (Magnoliales; Canellaceae). Nucleic Acids Research, 20,6101-6102.
DOI URL PMID |
| [30] | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 24,4876-4882. |
| [31] |
Wei XX, Wang XQ (2004) Recolonization and radiation in Larix (Pinaceae): evidence from nuclear ribosomal DNA paralogues. Molecular Ecology, 13,3115-3123.
DOI URL PMID |
| [32] |
Wei XX, Wang XQ, Hong DY (2003) Marked intragenomic heterogeneity and geographical differentiation of nrDNA ITS in Larix potaninii (Pinaceae). Journal of Molecular Evolution, 57,623-635.
URL PMID |
| [33] | Whitelock LM (2002) The Cycads. Timber Press, Inc.,Portland. |
| [34] |
Won H, Renner SS (2005) The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Molecular Phylogenetics and Evolution, 36,581-597.
DOI URL PMID |
| [35] |
Zheng XY, Cai DY, Yao LH, Teng YW (2008) Non-concerted ITS evolution, early origin and phylogenetic utility of ITS pseudogenes in Pyrus. Molecular Phylogenetics and Evolution, 48,892-903.
URL PMID |
| [36] |
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31,3406-3415.
DOI URL PMID |
| [1] | Gang Ren, En Li, Shiye Zhao, Yanqiong Jiang, Shasha Wang, Sixian Tang, Huijian Hu. Correlation between color polymorphism and the MC1R gene of Lanius schach [J]. Biodiv Sci, 2020, 28(6): 688-694. |
| [2] | Hongzheng Ma, Shanshan Li, Song Ge, Silan Dai, Wenli Chen. Isolation of SSR markers for two related second-generation energy crop species, Miscanthus nepalensis and M. nudipes (Poaceae) [J]. Biodiv Sci, 2011, 19(5): 535-542. |
| [3] | Lichuan Dai, Minglong Zhang, Jiye Liu, Xiaobai Li, Hairui Cui. Genetic diversity in Chinese rapeseed (Brassica napus) cultivars based on EST-SSR markers [J]. Biodiv Sci, 2009, 17(5): 482-489. |
| [4] | Liying Geng, Chuansheng Zhang, Lixin Du. Single nucleotide polymorphisms in chicken genomic pre-microRNA [J]. Biodiv Sci, 2009, 17(3): 248-256. |
| [5] | Zhao-E Pan, Junling Sun, Yinhua Jia, Zhongli Zhou, Baoyin Pang, Xiongming Du. Screening of SSR core primers with polymorphism on a cotton panel [J]. Biodiv Sci, 2008, 16(6): 555-561. |
| [6] | Lin Wei, Shenggui Liu, Xianwei Shi. Genetic diversity of the Xuefeng black bone chicken based on microsatellite markers [J]. Biodiv Sci, 2008, 16(5): 503-508. |
| [7] | Zhijun Dong, Hui Huang, Liangmin Huang, Yuanchao Li, Xiubao Li. PCR-RFLP analysis of large subunit rDNA of symbiotic dinoflagellates in scleractinian corals from Luhuitou fringing reef of Sanya, Hainan [J]. Biodiv Sci, 2008, 16(5): 498-502. |
| [8] | Yousheng Rao, Xiquan Zhang. Copy-number variant: new annotation for genetic variation across genomes [J]. Biodiv Sci, 2008, 16(4): 399-406. |
| [9] | Xiaofeng Zhao, Junhong Wu, Ningying Xu, Xiaoxiang Hu, Ning Li. Genetic structure of Jinhua pig and genetic differentiation of Jinhua pig and Taihu pig breeds based on microsatellite DNA markers [J]. Biodiv Sci, 2008, 16(4): 339-345. |
| [10] | Jianmin Wang, Wenbin Yue. Genetic relationships of domestic sheep and goats in the lower reaches of the Yellow River based on microsatellite analysis [J]. Biodiv Sci, 2008, 16(1): 53-62. |
| [11] | Hongfei Wang, Yan Wei. Seed polymorphism and fruit-set patterns of Salsola affinis [J]. Biodiv Sci, 2007, 15(4): 419-424. |
| [12] | Tian Zhang, Zuozhou Li, Yaling Liu, Zhengwang Jiang, Hongwen Huang . Genetic diversity, gene introgression and homoplasy in sympatric popu-lations of the genus Actinidia as revealed by chloroplast microsatellite markers [J]. Biodiv Sci, 2007, 15(1): 1-22. |
| [13] | Yunjie Tu, Kuanwei Chen, Qingping Tang, Jinyu Wang, Yushi Gao, Rong Gu, Qinglian Ge. Genetic diversity of 13 indigenous grey goose breeds in China based on microsatellite markers [J]. Biodiv Sci, 2006, 14(2): 152-158. |
| [14] | HUANG Bo, WANG Cheng-Shu, WANG Bin, FAN Mei-Zhen, LI Zeng-Zhi. nrDNA ITS analysis of Paecilomyces farinosus isolates [J]. Biodiv Sci, 2003, 11(6): 480-485. |
| [15] | ZOU Yu-Ping, GE Song. A novel molecular marker—SNPs and its application [J]. Biodiv Sci, 2003, 11(5): 370-382. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()