



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (7): 24475. DOI: 10.17520/biods.2024475 cstr: 32101.14.biods.2024475
• Original Papers: Plant Diversity • Previous Articles Next Articles
Deju Yu1,2, Yunyun He1,2( ), Min Cao1(
), Min Cao1( ), Gang Wang1(
), Gang Wang1( ), Jie Yang1,*(
), Jie Yang1,*( )(
)( )
)
Received:2024-10-31
Accepted:2025-03-11
Online:2025-07-20
Published:2025-08-06
Contact:
*E-mail: yangjie@xtbg.org.cn
Supported by:Deju Yu, Yunyun He, Min Cao, Gang Wang, Jie Yang. Coexistence mechanism of tropical forest tree species based on metabolomics and transcriptomics technologies: Taking Ficus species as an example[J]. Biodiv Sci, 2025, 33(7): 24475.
| 编号 Code | 物种 Species | 多度 Abundance | 生境 Habitat |
|---|---|---|---|
| 1 | 青藤公 Ficus langkokensis (FICULA) | 1,412 | 半坡 Slope |
| 2 | 水同木 Ficus fistulosa (FICUFI) | 917 | 半坡 Slope |
| 3 | 对叶榕 Ficus hispida (FICUHI) | 501 | 半坡 Slope |
| 4 | 北碚榕 Ficus beipeiensis (FICUBEI) | 214 | 沟谷 Valley |
| 5 | 杂色榕 Ficus variegate (FICUVAR) | 172 | 半坡 Slope |
| 6 | 大果榕 Ficus auriculata (FICUAU) | 105 | 沟谷 Valley |
| 7 | 金毛榕 Ficus fulva (FICUFU) | 103 | 半坡 Slope |
| 8 | 大叶水榕 Ficus glaberrima (FICUGL) | 79 | 沟谷 Valley |
| 9 | 黄毛榕 Ficus esquiroliana (FICUES) | 79 | 半坡 Slope |
| 10 | 鸡嗉子榕 Ficus semicordata (FICUSE) | 39 | 半坡 Slope |
| 11 | 棒果榕 Ficus subincisa (FICUSU) | 31 | 沟谷 Valley |
| 12 | 粗叶榕 Ficus altissima (FICUAL) | 30 | 沟谷 Valley |
| 13 | 苹果榕 Ficus oligodon (FICUOL) | 30 | 沟谷 Valley |
Table 1 Basic information of 13 target Ficus species within the 20-ha dynamics plot in Xishuangbanna tropical seasonal rainforest
| 编号 Code | 物种 Species | 多度 Abundance | 生境 Habitat |
|---|---|---|---|
| 1 | 青藤公 Ficus langkokensis (FICULA) | 1,412 | 半坡 Slope |
| 2 | 水同木 Ficus fistulosa (FICUFI) | 917 | 半坡 Slope |
| 3 | 对叶榕 Ficus hispida (FICUHI) | 501 | 半坡 Slope |
| 4 | 北碚榕 Ficus beipeiensis (FICUBEI) | 214 | 沟谷 Valley |
| 5 | 杂色榕 Ficus variegate (FICUVAR) | 172 | 半坡 Slope |
| 6 | 大果榕 Ficus auriculata (FICUAU) | 105 | 沟谷 Valley |
| 7 | 金毛榕 Ficus fulva (FICUFU) | 103 | 半坡 Slope |
| 8 | 大叶水榕 Ficus glaberrima (FICUGL) | 79 | 沟谷 Valley |
| 9 | 黄毛榕 Ficus esquiroliana (FICUES) | 79 | 半坡 Slope |
| 10 | 鸡嗉子榕 Ficus semicordata (FICUSE) | 39 | 半坡 Slope |
| 11 | 棒果榕 Ficus subincisa (FICUSU) | 31 | 沟谷 Valley |
| 12 | 粗叶榕 Ficus altissima (FICUAL) | 30 | 沟谷 Valley |
| 13 | 苹果榕 Ficus oligodon (FICUOL) | 30 | 沟谷 Valley |
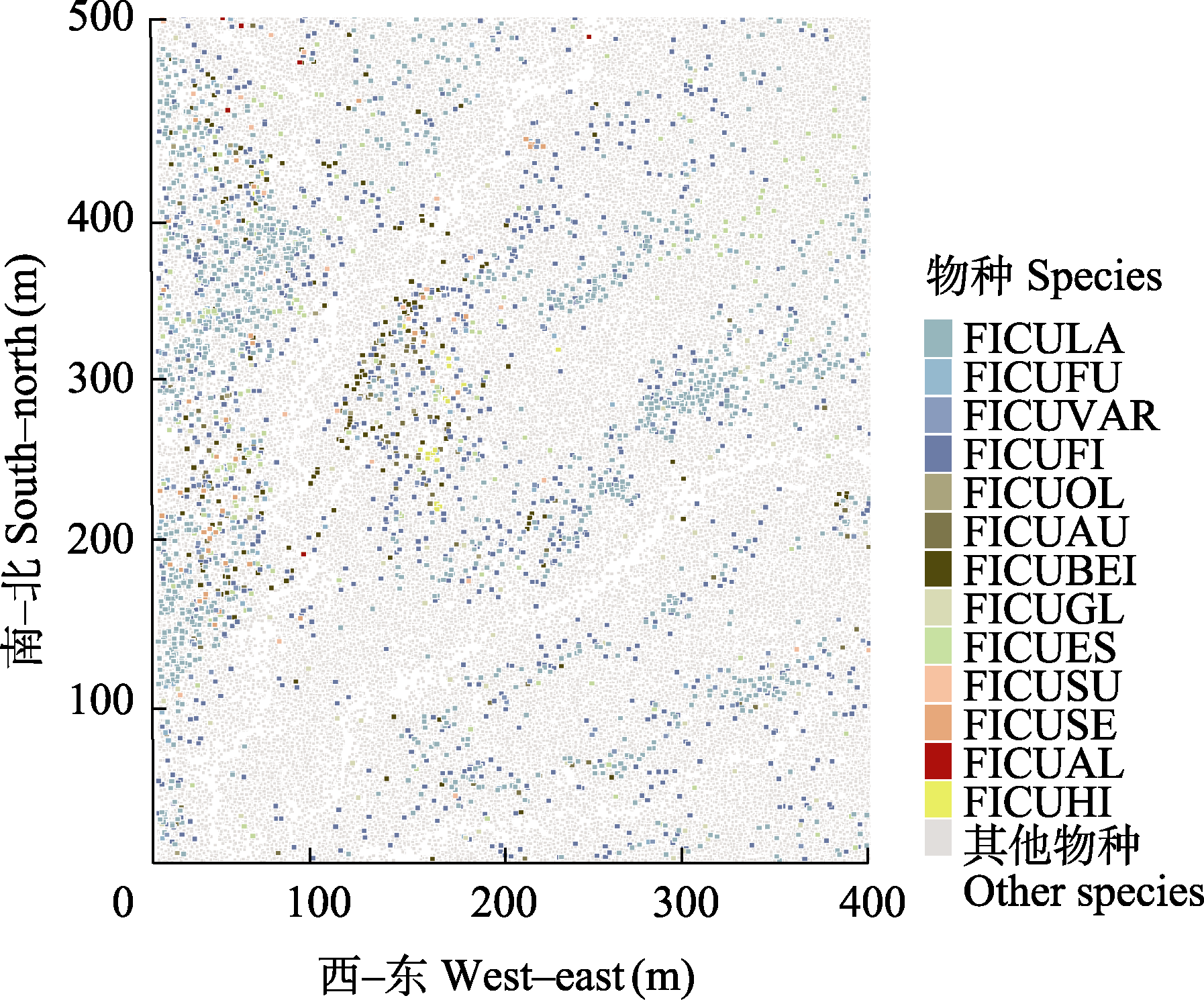
Fig. 1 Spatial distribution of 13 Ficus species within the 20-ha dynamics plot in Xishuangbanna tropical seasonal rainforest (species abbreviations see Table 1)
| 性状 Trait | K值 K value | P值 P value |
|---|---|---|
| 物理防御性状 Physical defense traits | ||
| 叶片厚度 Leaf thickness (mm) | 0.280 | 0.967 |
| 叶片韧性 Leaf toughness | 0.416 | 0.475 |
| 叶干物质含量 Leaf dry matter content (LDMC) (mg/g) | 0.270 | 0.920 |
| 叶片碳氮比 C : N | 1.063 | 0.008* |
| 防御基因 Defense genes | 0.216 | 0.992 |
| 化学防御性状 Chemical defense traits | ||
| 总代谢物 All plants secondary metabolites | 0.609 | 0.154 |
| 生物碱类 Alkaloids | 0.286 | 0.826 |
| 氨基酸/肽类 Amino acids/peptides | 0.627 | 0.090 |
| 碳水化合物类 Carbohydrates | 0.623 | 0.144 |
| 脂肪酸类 Fatty acids | 0.727 | 0.136 |
| 聚酮类 Polyketides | 0.647 | 0.102 |
| 莽草酸-苯丙素类 Shikimates and phenylpropanoids | 0.840 | 0.074 |
| 萜类 Terpenoids | 0.433 | 0.420 |
Table 2 Phylogenetic signal tests for physical defense traits, defense-related genes, and chemical defense traits of 13 Ficus species within the 20-ha dynamics plot in Xishuangbanna tropical seasonal rainforest
| 性状 Trait | K值 K value | P值 P value |
|---|---|---|
| 物理防御性状 Physical defense traits | ||
| 叶片厚度 Leaf thickness (mm) | 0.280 | 0.967 |
| 叶片韧性 Leaf toughness | 0.416 | 0.475 |
| 叶干物质含量 Leaf dry matter content (LDMC) (mg/g) | 0.270 | 0.920 |
| 叶片碳氮比 C : N | 1.063 | 0.008* |
| 防御基因 Defense genes | 0.216 | 0.992 |
| 化学防御性状 Chemical defense traits | ||
| 总代谢物 All plants secondary metabolites | 0.609 | 0.154 |
| 生物碱类 Alkaloids | 0.286 | 0.826 |
| 氨基酸/肽类 Amino acids/peptides | 0.627 | 0.090 |
| 碳水化合物类 Carbohydrates | 0.623 | 0.144 |
| 脂肪酸类 Fatty acids | 0.727 | 0.136 |
| 聚酮类 Polyketides | 0.647 | 0.102 |
| 莽草酸-苯丙素类 Shikimates and phenylpropanoids | 0.840 | 0.074 |
| 萜类 Terpenoids | 0.433 | 0.420 |
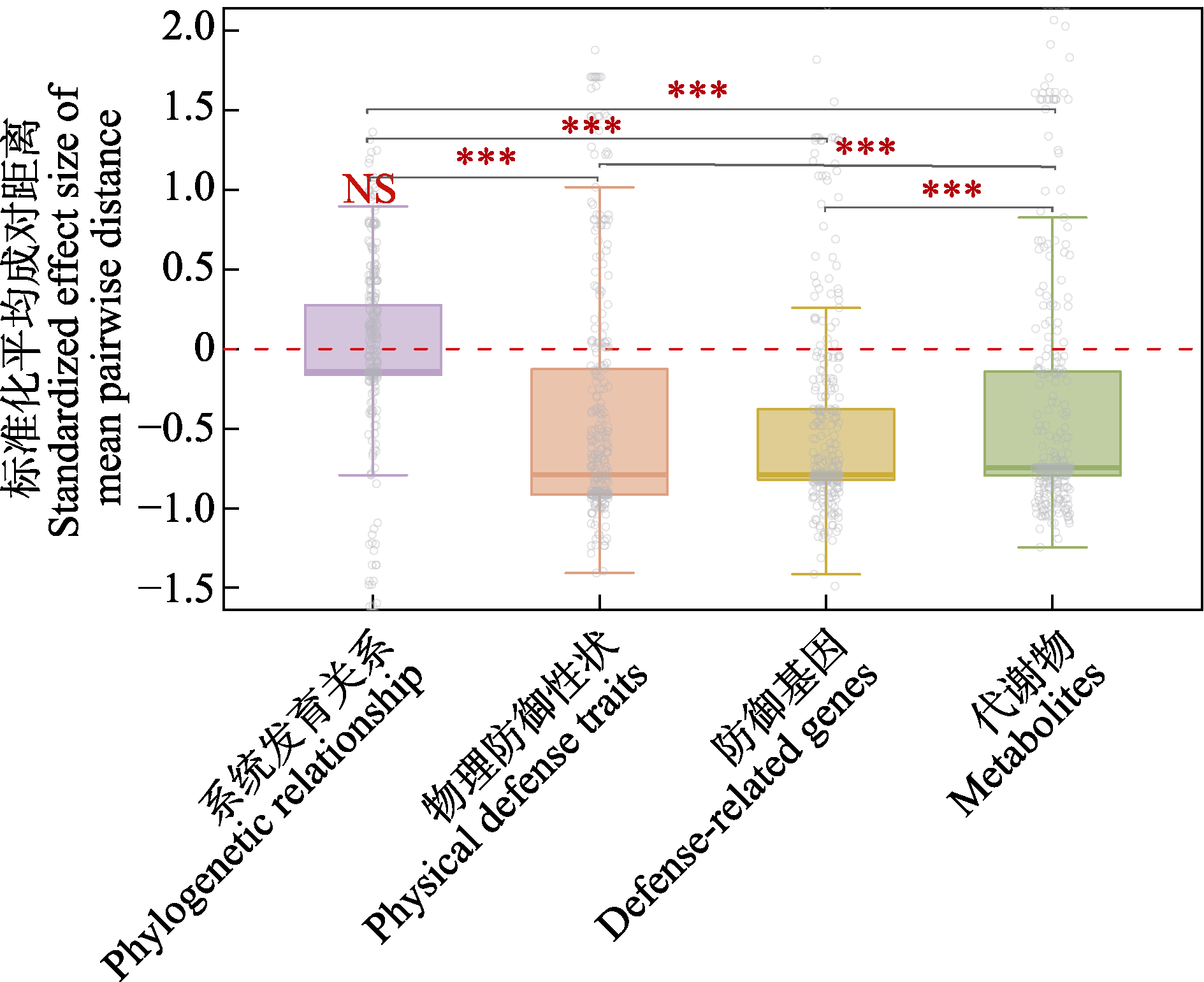
Fig. 2 Standardized effect size of mean pairwise distance for phylogenetic relationships, physical defense traits, defense genes, and metabolites among Ficus species within the 20-ha dynamics plot in Xishuangbanna tropical seasonal rainforest (*** indicates P < 0.01 for comparisons between groups; NS indicates P > 0.05 for comparisons with zero)
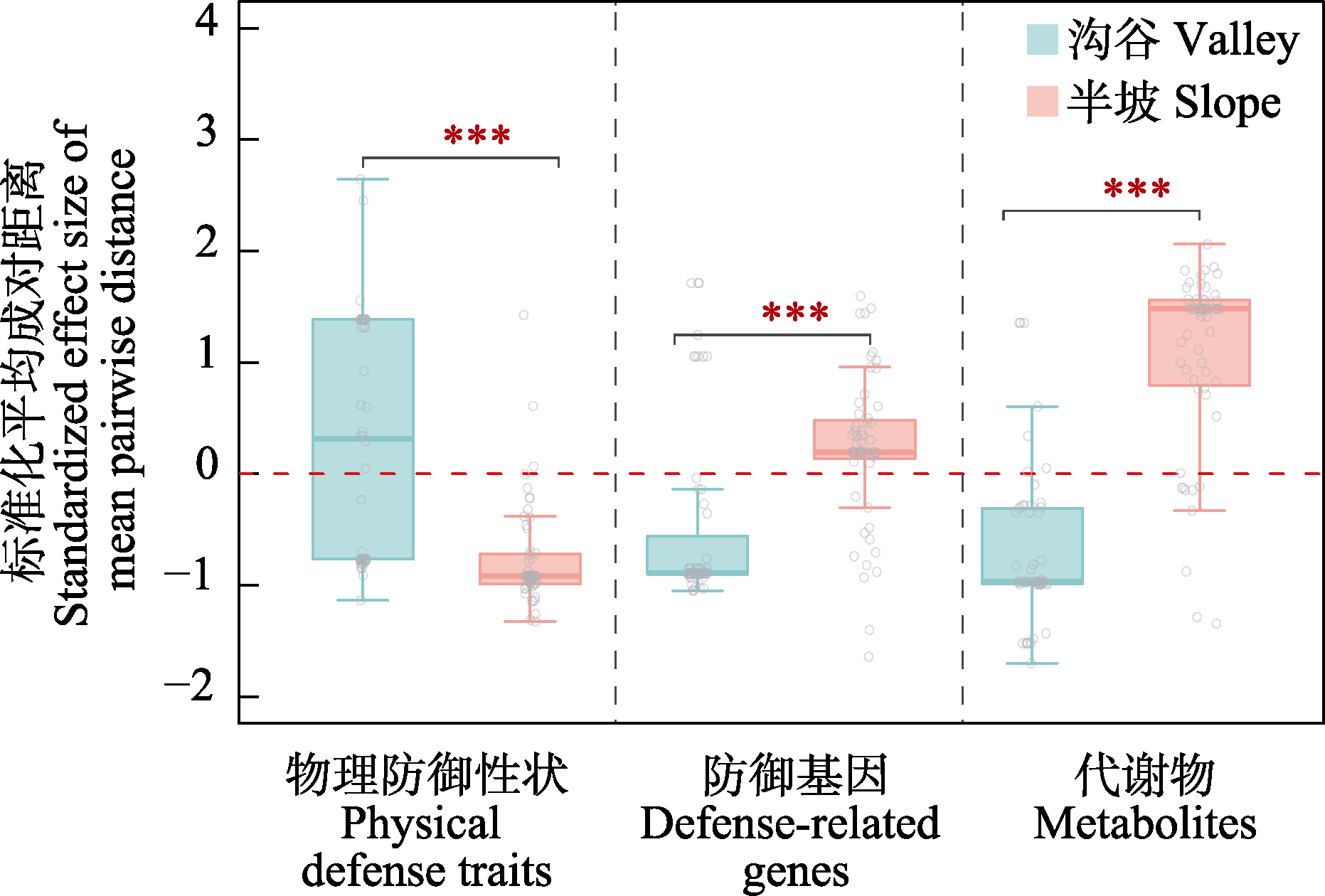
Fig. 3 Standardized effect size of mean pairwise distance of defensive traits in Ficus species from different habitats within the 20-ha dynamics plot in Xishuangbanna tropical seasonal rainforest (*** indicates P < 0.01 for comparisons between groups)
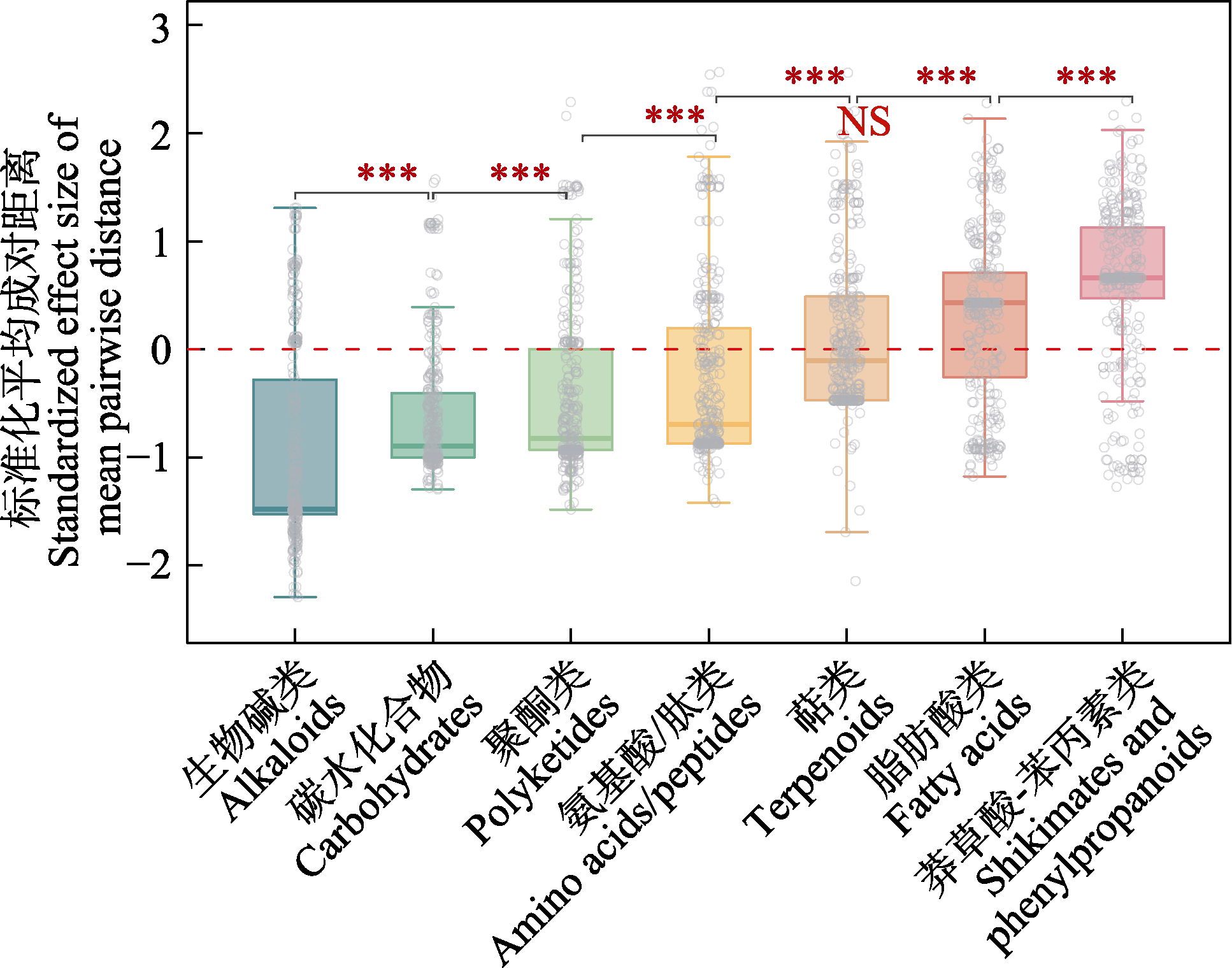
Fig. 4 Standardized effect size of mean pairwise distance for different metabolites among Ficus species within the 20-ha dynamics plot in Xishuangbanna tropical seasonal rainforest (*** indicates P < 0.01 for comparisons between groups; NS indicates P > 0.05 for comparisons with zero)
| [1] |
Adler PB, HilleRisLambers J, Levine JM (2007) A niche for neutrality. Ecology Letters, 10, 95-104.
PMID |
| [2] | Andrews S, Krueger F, Segonds-Pichon A, Biggins L, Krueger C, Wingett S (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. (accessed on 2024-06-11) |
| [3] | Asefa M, Song XY, Cao M, Lasky JR, Yang J (2021) Temporal trait plasticity predicts the growth of tropical trees. Journal of Vegetation Science, 32, e013056. |
| [4] | Ashton PS (1969) Speciation among tropical forest trees: Some deductions in the light of recent evidence. Biological Journal of the Linnean Society, 1, 155-196. |
| [5] | Bézier A, Lambert B, Baillieul F (2002) Study of defense-related gene expression in grapevine leaves and berries infected with Botrytis cinerea. European Journal of Plant Pathology, 108, 111-120. |
| [6] |
Blomberg SP, Garland JT, Ives AR (2003) Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution, 57, 717-745.
DOI PMID |
| [7] |
Böcker S, Letzel MC, Lipták Z, Pervukhin A (2009) SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics, 25, 218-224.
DOI PMID |
| [8] |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114-2120.
DOI PMID |
| [9] | Cao M, Zou XM, Warren M, Zhu H (2006) Tropical forests of Xishuangbanna, China. Biotropica, 38, 306-309. |
| [10] |
Chung M, Bruno VM, Rasko DA, Cuomo CA, Muñoz JF, Livny J, Shetty AC, Mahurkar A, Dunning Hotopp JC (2021) Best practices on the differential expression analysis of multi-species RNA-seq. Genome Biology, 22, 121.
DOI PMID |
| [11] | Cornelissen JHC, Lavorel S, Garnier E, Díaz S, Buchmann N, Gurvich DE, Reich PB, ter Steege H, Morgan HD, van der Heijden MGA, Pausas JG, Poorter H (2003) A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Australian Journal of Botany, 51, 335-380. |
| [12] |
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772.
DOI PMID |
| [13] | Davies RG, Orme CDL, Webster AJ, Jones KE, Blackburn TM, Gaston KJ (2007) Environmental predictors of global parrot (Aves: Psittaciformes) species richness and phylogenetic diversity. Global Ecology and Biogeography, 16, 220-233. |
| [14] | Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11-15. |
| [15] | Endara MJ, Coley PD, Ghabash G, Nicholls JA, Dexter KG, Donoso DA, Stone GN, Pennington RT, Kursar TA (2017) Coevolutionary arms race versus host defense chase in a tropical herbivore-plant system. Proceedings of the National Academy of Sciences, USA, 114, E7499-E7505. |
| [16] |
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics, 28, 3150-3152.
DOI PMID |
| [17] |
Gause GF (1934) Experimental analysis of Vito Volterra’s mathematical theory of the struggle for existence. Science, 79, 16-17.
PMID |
| [18] | Gershenzon J, Ullah C (2022) Plants protect themselves from herbivores by optimizing the distribution of chemical defenses. Proceedings of the National Academy of Sciences, USA, 119, e2120277119. |
| [19] |
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29, 644-652.
DOI PMID |
| [20] |
Haak DC, Ballenger BA, Moyle LC (2014) No evidence for phylogenetic constraint on natural defense evolution among wild tomatoes. Ecology, 95, 1633-1641.
PMID |
| [21] | Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols, 8, 1494-1512. |
| [22] | Harrison RD (2005) Figs and the diversity of tropical rainforests. BioScience, 55, 1053-1064. |
| [23] | Harvey PH, Pagel M (1991) The Comparative Method in Evolutionary Biology. Oxford University Press, Oxford. |
| [24] | Hutchinson GE (1961) The paradox of the plankton. The American Naturalist, 95, 137-145. |
| [25] | Jin L, Liu JJ, Xiao TW, Li QM, Lin LX, Shao XN, Ma CX, Li BH, Mi XC, Ren HB, Qiao XJ, Lian JY, Hao G, Ge XJ (2022) Plastome‐based phylogeny improves community phylogenetics of subtropical forests in China. Molecular Ecology Resources, 22, 319-333. |
| [26] |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772-780.
DOI PMID |
| [27] |
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26, 1463-1464.
DOI PMID |
| [28] |
Kim HW, Wang M, Leber CA, Nothias LF, Reher R, Kang KB, van der Hooft JJJ, Dorrestein PC, Gerwick WH, Cottrell GW (2021) NPClassifier: A deep neural network-based structural classification tool for natural products. Journal of Natural Products, 84, 2795-2807.
DOI PMID |
| [29] |
Kristiansson E, Österlund T, Gunnarsson L, Arne G, Joakim Larsson DG, Nerman O (2013) A novel method for cross-species gene expression analysis. BMC Bioinformatics, 14, 70.
DOI PMID |
| [30] | Kursar TA, Dexter KG, Lokvam J, Pennington RT, Richardson J E, Weber MG, Coley PD (2009) The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proceedings of the National Academy of Sciences, USA, 106, 18073-18078. |
| [31] | Lan G, Zhu H, Cao M, Hu Y, Wang H, Deng X, Zhou S, Cui J, Huang J, He Y, Liu L, Xu H, Song J (2009) Spatial dispersion patterns of trees in a tropical rainforest in Xishuangbanna, Southwest China. Ecological Research, 24, 1117-1124. |
| [32] | Larget BR, Kotha SK, Dewey CN, Ané C (2010) BUCKy: Gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics, 26, 2910-2911. |
| [33] | Lasky JR, Yang J, Zhang G, Cao M, Tang Y, Keitt TH (2014) The role of functional traits and individual variation in the co-occurrence of Ficus species. Ecology, 95, 978-990. |
| [34] | Malhi Y (2012) The productivity, metabolism and carbon cycle of tropical forest vegetation. Journal of Ecology, 100, 65-75. |
| [35] |
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20, 1297-1303.
DOI PMID |
| [36] | Mirarab S, Warnow T (2015) ASTRAL-II: Coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics, 31, i44-i52. |
| [37] | Newman A (2002) Tropical Rainforest:Our Most Valuable and Endangered Habitat with a Blueprint for Its Survival into the Third Millennium. Checkmark Books, New York. |
| [38] | R Core Team (2023) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. https://www.R-project.org/. (accessed on 2024-06-11) |
| [39] | Price MN, Dehal PS, Arkin AP (2010) FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE, 5, e9490. |
| [40] | Richards LA, Dyer LA, Forister ML, Smilanich AM, Dodson CD, Leonard MD, Jeffrey CS (2015) Phytochemical diversity drives plant-insect community diversity. Proceedings of the National Academy of Sciences, USA, 112, 10973-10978. |
| [41] |
Sedio BE, Rojas Echeverri JC, Boya PCA, Wright SJ (2017) Sources of variation in foliar secondary chemistry in a tropical forest tree community. Ecology, 98, 616-623.
DOI PMID |
| [42] | Seelig H, Stoner RJ, Linden JC (2012) Irrigation control of cowpea plants using the measurement of leaf thickness under greenhouse conditions. Irrigation Science, 30, 247-257. |
| [43] | Seibold SD, Cadotte MW, MacIvor JS, Thorn S, Müller J (2018) The necessity of multitrophic approaches in community ecology. Trends in Ecology & Evolution, 33, 754-764. |
| [44] |
Shafer MER (2019) Cross-species analysis of single-cell transcriptomic data. Frontiers in Cell and Developmental Biology, 7, 175.
DOI PMID |
| [45] |
Smith SA, Dunn CW (2008) Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics, 24, 715-716.
DOI PMID |
| [46] |
Solís-Lemus C, Bastide P, Ané C (2017) PhyloNetworks: A package for phylogenetic networks. Molecular Biology and Evolution, 34, 3292-3298.
DOI PMID |
| [47] | Spasojevic MJ, Suding KN (2012) Inferring community assembly mechanisms from functional diversity patterns: The importance of multiple assembly processes. Journal of Ecology, 100, 652-661. |
| [48] |
Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688-2690.
DOI PMID |
| [49] | Su ZH, Sasaki A, Kusumi J, Chou PA, Tzeng HY, Li HQ, Yu H (2022) Pollinator sharing, copollination, and speciation by host shifting among six closely related dioecious fig species. Communications Biology, 5, 284. |
| [50] | Swenson NG (2013) The assembly of tropical tree communities—The advances and shortcomings of phylogenetic and functional trait analyses. Ecography, 36, 264-276. |
| [51] | Swenson NG (2014) Functional and Phylogenetic Ecology in R. Springer, New York. |
| [52] | Swenson NG, Iida Y, Howe R, Wolf A, Umaña MN, Petprakob K, Turner BL, Ma KP (2017) Tree co-occurrence and transcriptomic response to drought. Nat‘ure Communications, 8, 1996. |
| [53] | Swenson NG, Rubio VE (2024) The functional underpinnings of tropical forest dynamics—Functional traits, groups, and unmeasured diversity. Biotropica, 57, e13360. |
| [54] |
Tissier A (2018) Plant secretory structures: More than just reaction bags. Current Opinion in Biotechnology, 49, 73-79.
DOI PMID |
| [55] |
Valladares F, Bastias CC, Godoy O, Granda E, Escudero A (2015) Species coexistence in a changing world. Frontiers in Plant Science, 6, 866.
DOI PMID |
| [56] | van Dongen S (2000) Graph Clustering by Flow Simulation. PhD dissertation. University of Utrecht, Utrecht. |
| [57] |
Wang G, Zhang XT, Herre EA, McKey D, Machado CA, Yu WB, Cannon CH, Arnold ML, Pereira RAS, Ming R, Liu YF, Wang YB, Ma DN, Chen J (2021) Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig-wasp pollination mutualism. Nature Communications, 12, 718.
DOI PMID |
| [58] | Wang MX, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu WT, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu CC, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw CC, Yang YL, Humpf HU, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, Boya PCA, Torres-Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O’Neill EC, Briand E, Helfrich EJN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodríguez AMC, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard PM, Phapale P, Nothias LF, Alexandrov T, Litaudon M, Wolfender JL, Kyle JE, Metz TO, Peryea T, Nguyen DT, VanLeer D, Shinn P, Jadhav A, Müller R, Waters KM, Shi WY, Liu XT, Zhang LX, Knight R, Jensen PR, Palsson BØ, Pogliano K, Linington RG, Gutiérrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC, Bandeira N (2016) Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology, 34, 828-837. |
| [59] | Wang XZ (2022) Study on the Coexistence of Community Tree Species Based on Secondary Metabolites: A Case Study of Xishuangbanna Tropical Seasonal Rain Forest. PhD dissertation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming. (in Chinese with English abstract) |
| [王雪昭 (2022) 基于次生代谢产物的群落物种共存研究——以西双版纳热带季节雨林为例. 博士学位论文, 中国科学院西双版纳热带植物园, 昆明.] | |
| [60] |
Wang XZ, He YY, Sedio BE, Jin L, Ge XJ, Glomglieng S, Cao M, Yang JH, Swenson NG, Yang J (2023) Phytochemical diversity impacts herbivory in a tropical rainforest tree community. Ecology Letters, 26, 1898-1910.
DOI PMID |
| [61] | Wang XZ, Sun SW, Sedio BE, Glomglieng S, Cao M, Cao KF, Yang JH, Zhang JL, Yang J (2022) Niche differentiation along multiple functional-trait dimensions contributes to high local diversity of Euphorbiaceae in a tropical tree assemblage. Journal of Ecology, 110, 2731-2744. |
| [62] |
Wang YT, Yang Y, Sun XL, Ji J (2023) Development of a widely-targeted metabolomics method based on gas chromatography-mass spectrometry. Chinese Journal of Chromatography, 41, 520-526. (in Chinese with English abstract)
DOI |
|
[王亚婷, 杨阳, 孙秀兰, 纪剑 (2023) 基于气相色谱-质谱法的广泛靶向代谢组学方法开发. 色谱, 41, 520-526.]
DOI |
|
| [63] |
Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM (2018) BUSCO applications from quality assessments to gene prediction and phylogenomics. Molecular Biology and Evolution, 35, 543-548.
DOI PMID |
| [64] | Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Annual Review of Ecology, Evolution, and Systematics, 33, 475-505. |
| [65] | Webb CO, Cannon CH, Davies SJ (2008) Ecological organization, biogeography, and the phylogenetic structure of tropical forest tree communities. Tropical Forest Community Ecology, 79-97. |
| [66] |
Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, Damschen EI, Jonathan Davies T, Grytnes JA, Harrison SP, Hawkins BA, Holt RD, McCain CM, Stephens PR (2010) Niche conservatism as an emerging principle in ecology and conservation biology. Ecology Letters, 13, 1310-1324.
DOI PMID |
| [67] |
Wink M (2018) Plant secondary metabolites modulate insect behavior-steps toward addiction? Frontiers in Physiology, 9, 364.
DOI PMID |
| [68] | Yang J (2013) Disentangling Mechanisms Underlying Tree Species Coexistence: Integrating Phylogenetic and Functional Dimensions. PhD dissertation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming. (in Chinese with English abstract) |
| [杨洁 (2013) 系统发育和功能性状维度的树种共存机制研究. 博士学位论文, 中国科学院西双版纳热带植物园, 昆明.] | |
| [69] | Yang J, Fan H, He YY, Wang G, Cao M, Swenson NG (2024) Functional genomics and co-occurrence in a diverse tropical tree genus: The roles of drought- and defence-related genes. Journal of Ecology, 112, 575-589. |
| [70] | Yang J, Zhang G, Ci X, Swenson NG, Cao M, Sha L, Li J, Baskin CC, Ferry Slik JW, Lin LX (2014) Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Functional Ecology, 28, 520-529. |
| [71] | Zambrano J, Iida Y, Howe R, Lin LX, Umana MN, Wolf A, Worthy SJ, Swenson NG (2017) Neighbourhood defence gene similarity effects on tree performance: A community transcriptomic approach. Journal of Ecology, 105, 616-626. |
| [72] | Zhao BG (1999) Nematicidal activity of quinolizidine alkaloids and the functional group pairs in their molecular structure. Journal of Chemical Ecology, 25, 2205-2214. |
| [73] | Zhao J, Segar ST, McKey D, Chen J (2021) Macroevolution of defense syndromes in Ficus (Moraceae). Ecological Monographs, 91, e01428. |
| [1] | Hong Deyuan. A brief discussion on methodology in taxonomy [J]. Biodiv Sci, 2025, 33(2): 24541-. |
| [2] | Yajun Sun. What do higher or lower organisms mean—Clarify the meaning and validity of the biological ladder implied by On the Origin of Species [J]. Biodiv Sci, 2025, 33(1): 24394-. |
| [3] | Chuan Jin, Zijia Zhang, Kai Di, Weirong Zhang, Dong Qiao, Siyuan Cheng, Zhongmin Hu. A dataset on fluorescence, photosynthesis gas exchange, and leaf traits of Hainan tropical rainforest plant species [J]. Biodiv Sci, 2024, 32(9): 24139-. |
| [4] | Fei Duan, Mingzhang Liu, Hongliang Bu, Le Yu, Sheng Li. Effects of urbanization on bird community composition and functional traits: A case study of the Beijing-Tianjin-Hebei region [J]. Biodiv Sci, 2024, 32(8): 23473-. |
| [5] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [6] | Yanyu Ai, Haixia Hu, Ting Shen, Yuxuan Mo, Jinhua Qi, Liang Song. Vascular epiphyte diversity and the correlation analysis with host tree characteristics: A case in a mid-mountain moist evergreen broad-leaved forest, Ailao Mountains [J]. Biodiv Sci, 2024, 32(5): 24072-. |
| [7] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [8] | Jialong Ren, Yongzhen Wang, Yilin Feng, Wenzhi Zhao, Qihan Yan, Chang Qin, Jing Fang, Weidong Xin, Jiliang Liu. Beetle data set collected by pitfall trapping in the gobi desert of the Hexi Corridor [J]. Biodiv Sci, 2024, 32(2): 23375-. |
| [9] | Jiayi Feng, Juyu Lian, Yujun Feng, Dongxu Zhang, Honglin Cao, Wanhui Ye. Effects of vertical stratification on community structure and functions in a subtropical, evergreen broad-leaved forest in the Dinghushan National Nature Reserve [J]. Biodiv Sci, 2024, 32(12): 24306-. |
| [10] | Chen-Kun Jiang, Wen-Bin Yu, Guang-Yuan Rao, Huaicheng Li, Julien B. Bachelier, Hartmut H. Hilger, Theodor C. H. Cole. Plant Phylogeny Posters—An educational project on plant diversity from an evolutionary perspective [J]. Biodiv Sci, 2024, 32(11): 24210-. |
| [11] | Zhizhong Li, Shuai Peng, Qingfeng Wang, Wei Li, Shichu Liang, Jinming Chen. Cryptic diversity of the genus Ottelia in China [J]. Biodiv Sci, 2023, 31(2): 22394-. |
| [12] | Huiyin Song, Zhengyu Hu, Guoxiang Liu. Assessing advances in taxonomic research on Chlorellaceae (Chlorophyta) [J]. Biodiv Sci, 2023, 31(2): 22083-. |
| [13] | Ting Wang, Jiangping Shu, Yufeng Gu, Yanqing Li, Tuo Yang, Zhoufeng Xu, Jianying Xiang, Xianchun Zhang, Yuehong Yan. Insight into the studies on diversity of lycophytes and ferns in China [J]. Biodiv Sci, 2022, 30(7): 22381-. |
| [14] | Jianming Wang, Mengjun Qu, Yin Wang, Yiming Feng, Bo Wu, Qi Lu, Nianpeng He, Jingwen Li. The drivers of plant taxonomic, functional, and phylogenetic β-diversity in the gobi desert of northern Qinghai-Tibet Plateau [J]. Biodiv Sci, 2022, 30(6): 21503-. |
| [15] | Tian Luo, Fangyuan Yu, Juyu Lian, Junjie Wang, Jian Shen, Zhifeng Wu, Wanhui Ye. Impact of canopy vertical height on leaf functional traits in a lower subtropical evergreen broad-leaved forest of Dinghushan [J]. Biodiv Sci, 2022, 30(5): 21414-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()