



Biodiv Sci ›› 2022, Vol. 30 ›› Issue (5): 21485. DOI: 10.17520/biods.2021485 cstr: 32101.14.biods.2021485
• Original Papers: Genetic Diversity • Previous Articles Next Articles
Jing Cui1, Mingfang Xu1, Qun Zhang1,*( ), Yao Li1, Xiaoshu Zeng1, Sha Li2,3
), Yao Li1, Xiaoshu Zeng1, Sha Li2,3
Received:2021-11-27
Accepted:2022-01-18
Online:2022-05-20
Published:2022-03-13
Contact:
Qun Zhang
Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers[J]. Biodiv Sci, 2022, 30(5): 21485.
| 海域 Sea region | 采样地点 Sample site | 样本数量 Sample size | 样本编号 Sample no. | 经纬度 Coordinates | 采样时间 Sampling date | 样本来源 Sample source |
|---|---|---|---|---|---|---|
| 中国黄海 Yellow Sea, China | 辽宁丹东 LNDD | 30 | LNDD 01-14 | 39.86° N, 124.16° E | 2008.10 | 朱叶等, |
| LNDD 15-30 | 39.86° N, 124.16° E | 2021.04 | 本研究 This study | |||
| 山东青岛 SDQD | 36 | SDLSh 01-14 | 36.07° N, 120.42° E | 2008.10 | 朱叶等, | |
| SDQD 01-22 | 36.07° N, 120.38° E | 2018.04 | 本研究 This study | |||
| 中国东海 East China Sea | 浙江舟山 ZJZS | 42 | ZJDSh 01-13 | 30.19° N, 122.17° E | 2008.11 | 朱叶等, |
| ZJZS 01-29 | 29.99° N, 122.20° E | 2021.05 | 本研究 This study | |||
| 浙江温州 ZJWZ | 14 | ZJWZ 01-14 | 28.02° N, 120.79° E | 2011.11 | 本研究 This study | |
| 中国南海 South China Sea | 广东饶平 GDRP | 23 | GDRP 01-16 | 23.62° N, 116.94° E | 2008.12 | 朱叶等, |
| GDNA 01-07 | 23.42° N, 117.02° E | 2021.04 | 本研究 This study | |||
| 广东碣石 GDJS | 20 | GDJS 01-20 | 22.81° N, 115.82° E | 2014.08 | 本研究 This study | |
| 日本濑户内海 Seto inland sea, Japan | 日本明石 JAP | 35 | JAP 01-35 | 34.64° N, 134.98° E | 2019.07 | 本研究 This study |
| 合计 Total | - | 200 | - | - | - | - |
Table 1 Sample information of Pleuronichthys corunutus in this study
| 海域 Sea region | 采样地点 Sample site | 样本数量 Sample size | 样本编号 Sample no. | 经纬度 Coordinates | 采样时间 Sampling date | 样本来源 Sample source |
|---|---|---|---|---|---|---|
| 中国黄海 Yellow Sea, China | 辽宁丹东 LNDD | 30 | LNDD 01-14 | 39.86° N, 124.16° E | 2008.10 | 朱叶等, |
| LNDD 15-30 | 39.86° N, 124.16° E | 2021.04 | 本研究 This study | |||
| 山东青岛 SDQD | 36 | SDLSh 01-14 | 36.07° N, 120.42° E | 2008.10 | 朱叶等, | |
| SDQD 01-22 | 36.07° N, 120.38° E | 2018.04 | 本研究 This study | |||
| 中国东海 East China Sea | 浙江舟山 ZJZS | 42 | ZJDSh 01-13 | 30.19° N, 122.17° E | 2008.11 | 朱叶等, |
| ZJZS 01-29 | 29.99° N, 122.20° E | 2021.05 | 本研究 This study | |||
| 浙江温州 ZJWZ | 14 | ZJWZ 01-14 | 28.02° N, 120.79° E | 2011.11 | 本研究 This study | |
| 中国南海 South China Sea | 广东饶平 GDRP | 23 | GDRP 01-16 | 23.62° N, 116.94° E | 2008.12 | 朱叶等, |
| GDNA 01-07 | 23.42° N, 117.02° E | 2021.04 | 本研究 This study | |||
| 广东碣石 GDJS | 20 | GDJS 01-20 | 22.81° N, 115.82° E | 2014.08 | 本研究 This study | |
| 日本濑户内海 Seto inland sea, Japan | 日本明石 JAP | 35 | JAP 01-35 | 34.64° N, 134.98° E | 2019.07 | 本研究 This study |
| 合计 Total | - | 200 | - | - | - | - |
| 海域 Sea | 群体 Population | 数量 Sample size | 变异位点数 Variable sites | 单倍型数 Nh Number of haplotypes | 单倍型多样性 Hd Haplotype diversity | 核苷酸多样性 π Nucleotide diversity |
|---|---|---|---|---|---|---|
| 中国 China | LNDD | 29 | 23 | 20 | 0.9606 | 0.0064 |
| SDQD | 36 | 21 | 27 | 0.9682 | 0.0063 | |
| ZJZS | 42 | 17 | 31 | 0.9826 | 0.0059 | |
| ZJWZ | 14 | 13 | 12 | 0.9780 | 0.0057 | |
| GDRP | 23 | 10 | 15 | 0.9486 | 0.0055 | |
| GDJS | 20 | 18 | 16 | 0.9632 | 0.0061 | |
| 日本 Japan | JAP | 35 | 20 | 23 | 0.9647 | 0.0064 |
| 总计 Total | - | 199 | 43 | 90 | 0.9699 | 0.0061 |
Table 2 Genetic diversity of Pleuronichthys cornutus in coastal water of China and Japan based on CR sequence analysis
| 海域 Sea | 群体 Population | 数量 Sample size | 变异位点数 Variable sites | 单倍型数 Nh Number of haplotypes | 单倍型多样性 Hd Haplotype diversity | 核苷酸多样性 π Nucleotide diversity |
|---|---|---|---|---|---|---|
| 中国 China | LNDD | 29 | 23 | 20 | 0.9606 | 0.0064 |
| SDQD | 36 | 21 | 27 | 0.9682 | 0.0063 | |
| ZJZS | 42 | 17 | 31 | 0.9826 | 0.0059 | |
| ZJWZ | 14 | 13 | 12 | 0.9780 | 0.0057 | |
| GDRP | 23 | 10 | 15 | 0.9486 | 0.0055 | |
| GDJS | 20 | 18 | 16 | 0.9632 | 0.0061 | |
| 日本 Japan | JAP | 35 | 20 | 23 | 0.9647 | 0.0064 |
| 总计 Total | - | 199 | 43 | 90 | 0.9699 | 0.0061 |
| 地点 Sites | JAP | LNDD | SDQD | ZJZS | ZJWZ | GDRP | GDJS |
|---|---|---|---|---|---|---|---|
| JAP | - | 44.6264 | ∞ | ∞ | ∞ | ∞ | 29.9692 |
| LNDD | 0.0111 | - | 109.1491 | 51.9659 | 26.7628 | 34.0782 | ∞ |
| SDQD | -0.0070 | 0.0046 | - | ∞ | 238.7344 | 229.9147 | 198.7032 |
| ZJZS | -0.0046 | 0.0095 | -0.0082 | - | ∞ | ∞ | 56.3828 |
| ZJWZ | -0.0197 | 0.0184 | 0.0021 | -0.0062 | - | ∞ | 31.4081 |
| GDRP | -0.0130 | 0.0145 | 0.0022 | -0.0188 | -0.0071 | - | 30.4981 |
| GDJS | 0.0164 | -0.0046 | 0.0025 | 0.0088 | 0.0157 | 0.0161 | - |
Table 3 Pairwise FST estimates (below diagonal) and Nm (upper diagonal) across sampling sites of Pleuronichthys cornutus based on CR sequences analysis. * P < 0.05; ∞, the value of Nm is negative.
| 地点 Sites | JAP | LNDD | SDQD | ZJZS | ZJWZ | GDRP | GDJS |
|---|---|---|---|---|---|---|---|
| JAP | - | 44.6264 | ∞ | ∞ | ∞ | ∞ | 29.9692 |
| LNDD | 0.0111 | - | 109.1491 | 51.9659 | 26.7628 | 34.0782 | ∞ |
| SDQD | -0.0070 | 0.0046 | - | ∞ | 238.7344 | 229.9147 | 198.7032 |
| ZJZS | -0.0046 | 0.0095 | -0.0082 | - | ∞ | ∞ | 56.3828 |
| ZJWZ | -0.0197 | 0.0184 | 0.0021 | -0.0062 | - | ∞ | 31.4081 |
| GDRP | -0.0130 | 0.0145 | 0.0022 | -0.0188 | -0.0071 | - | 30.4981 |
| GDJS | 0.0164 | -0.0046 | 0.0025 | 0.0088 | 0.0157 | 0.0161 | - |
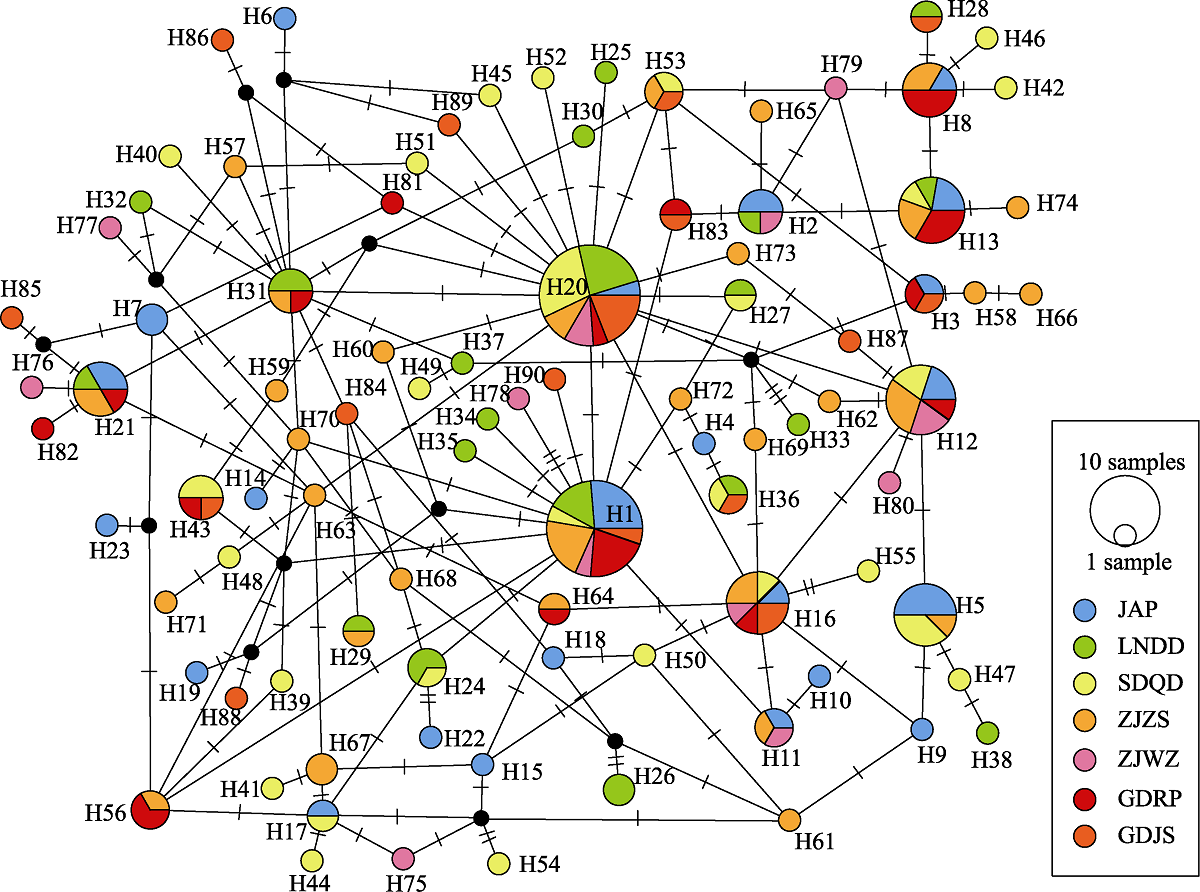
Fig. 1 Median-joining network of mtDNA CR haplotypes from 7 populations of Pleuronichthys cornutus. Different filling traits represent different populations; Node size corresponds to the haplotype frequencies; The node size of “1 sample” is 1 individual, and that of “10 samples” is 10 individuals; Black dots indicate haplotypes that theoretically present but not monitored; Short horizontal line on the line between the circles represents one mutation step. Location codes are the same as the sample sites in Table 1.
| 分组方式 Method of grouping | 变异比例 Percentage of variation (%) | FST 统计值 FST statistics (P-value) | ||
|---|---|---|---|---|
| 组群间 Among groups | 组群内群体间 Among populations within groups | 群体内 Within populations | ||
| 7个群体 Seven populations | 0.050 | - | 99.950 | 0.00054 (P > 0.05) |
| 中国6个群体和日本1个群体 Six populations from China and one population from Japan | -0.260 | 0.140 | 100.120 | -0.00118 (P > 0.05) |
| 濑户内海、中国黄海、东海和南海 Seto inland sea, Yellow Sea, East China Sea, and South China Sea | -0.410 | 0.410 | 100.000 | 0.00003 (P > 0.05) |
| 台湾海峡以南和台湾海峡以北 North of Taiwan Strait and south of Taiwan Strait | 0.990 | -0.160 | 99.170 | -0.00168 (P > 0.05) |
Table 4 AMOVA analysis of Pleuronichthys cornutus populations based on CR sequence
| 分组方式 Method of grouping | 变异比例 Percentage of variation (%) | FST 统计值 FST statistics (P-value) | ||
|---|---|---|---|---|
| 组群间 Among groups | 组群内群体间 Among populations within groups | 群体内 Within populations | ||
| 7个群体 Seven populations | 0.050 | - | 99.950 | 0.00054 (P > 0.05) |
| 中国6个群体和日本1个群体 Six populations from China and one population from Japan | -0.260 | 0.140 | 100.120 | -0.00118 (P > 0.05) |
| 濑户内海、中国黄海、东海和南海 Seto inland sea, Yellow Sea, East China Sea, and South China Sea | -0.410 | 0.410 | 100.000 | 0.00003 (P > 0.05) |
| 台湾海峡以南和台湾海峡以北 North of Taiwan Strait and south of Taiwan Strait | 0.990 | -0.160 | 99.170 | -0.00168 (P > 0.05) |
| 群体 Population | 数量 Sample size | CR/Cytb/ND2遗传多样性 CR/Cytb/ND2 genetic diversity | ||||
|---|---|---|---|---|---|---|
| 变异位点 Variable sites | 单倍型数 Nh Number of haplotypes | 单倍型多样性 Hd Haplotype diversity | 核苷酸多样性 π Nucleotide diversity | |||
| 中国 China | LNDD | 30 | 20/41/36 | 20/23/22 | 0.9563/0.9494/0.9678 | 0.0067/0.0048/0.0048 |
| SDQD | 30 | 19/44/47 | 23/27/26 | 0.9494/0.9908/0.9908 | 0.0063/0.0058/0.0053 | |
| ZJZS | 26 | 13/27/45 | 22/22/23 | 0.9877/0.9846/0.9815 | 0.0061/0.0039/0.0055 | |
| GDJS | 20 | 18/46/45 | 16/18/20 | 0.9632/0.9842/1.0000 | 0.0061/0.0063/0.0059 | |
| 日本 Japan | JAP | 34 | 20/40/32 | 22/25/20 | 0.9626/0.9768/0.9412 | 0.0063/0.0050/0.0041 |
| 合计 Total | 140 | 35/103/112 | 74/95/94 | 0.9683/0.9829/0.9811 | 0.0063/0.0051/0.0050 | |
Table 5 Sample information and genetic diversity indices of Pleuronichthys cornutus populations based on CR, Cytb, and ND2 sequences
| 群体 Population | 数量 Sample size | CR/Cytb/ND2遗传多样性 CR/Cytb/ND2 genetic diversity | ||||
|---|---|---|---|---|---|---|
| 变异位点 Variable sites | 单倍型数 Nh Number of haplotypes | 单倍型多样性 Hd Haplotype diversity | 核苷酸多样性 π Nucleotide diversity | |||
| 中国 China | LNDD | 30 | 20/41/36 | 20/23/22 | 0.9563/0.9494/0.9678 | 0.0067/0.0048/0.0048 |
| SDQD | 30 | 19/44/47 | 23/27/26 | 0.9494/0.9908/0.9908 | 0.0063/0.0058/0.0053 | |
| ZJZS | 26 | 13/27/45 | 22/22/23 | 0.9877/0.9846/0.9815 | 0.0061/0.0039/0.0055 | |
| GDJS | 20 | 18/46/45 | 16/18/20 | 0.9632/0.9842/1.0000 | 0.0061/0.0063/0.0059 | |
| 日本 Japan | JAP | 34 | 20/40/32 | 22/25/20 | 0.9626/0.9768/0.9412 | 0.0063/0.0050/0.0041 |
| 合计 Total | 140 | 35/103/112 | 74/95/94 | 0.9683/0.9829/0.9811 | 0.0063/0.0051/0.0050 | |
| 序列 Sequences | 地点 Sites | JAP | LNDD | SDQD | ZJZS | GDJS |
|---|---|---|---|---|---|---|
| CR | JAP | - | ∞ | ∞ | ∞ | 29.3686 |
| LNDD | -0.0051 | - | 77.9929 | 79.3722 | ∞ | |
| SDQD | -0.0060 | 0.0064 | - | ∞ | ∞ | |
| ZJZS | -0.0302 | 0.0063 | -0.0064 | - | 19.3728 | |
| GDJS | 0.0167 | -0.0036 | -0.0063 | 0.0252 | - | |
| Cytb | JAP | - | ∞ | ∞ | 133.9086 | 49.7008 |
| LNDD | -0.0059 | - | ∞ | ∞ | 53.3793 | |
| SDQD | -0.0001 | -0.0013 | - | 133.1898 | 111.8596 | |
| ZJZS | 0.0037 | -0.0091 | 0.0037 | - | 16.1058 | |
| GDJS | 0.0100 | 0.0093 | 0.0045 | 0.0301 | - | |
| ND2 | JAP | - | 164.5165 | 57.1701 | 369.8704 | 16.0618 |
| LNDD | 0.0030 | - | ∞ | 34.3918 | 26.5856 | |
| SDQD | 0.0087 | -0.0037 | - | 123.8781 | ∞ | |
| ZJZS | 0.0014 | 0.0143 | 0.0040 | - | 27.6373 | |
| GDJS | 0.0302* | 0.0185 | -0.0103 | 0.0178 | - |
Table 6 Pairwise FST estimates (below diagonal) and Nm (upper diagonal) between sampling sites of Pleuronichthys cornutus based on CR, Cytb and ND2 sequences. * P < 0.05; ∞, the value of Nm is negative.
| 序列 Sequences | 地点 Sites | JAP | LNDD | SDQD | ZJZS | GDJS |
|---|---|---|---|---|---|---|
| CR | JAP | - | ∞ | ∞ | ∞ | 29.3686 |
| LNDD | -0.0051 | - | 77.9929 | 79.3722 | ∞ | |
| SDQD | -0.0060 | 0.0064 | - | ∞ | ∞ | |
| ZJZS | -0.0302 | 0.0063 | -0.0064 | - | 19.3728 | |
| GDJS | 0.0167 | -0.0036 | -0.0063 | 0.0252 | - | |
| Cytb | JAP | - | ∞ | ∞ | 133.9086 | 49.7008 |
| LNDD | -0.0059 | - | ∞ | ∞ | 53.3793 | |
| SDQD | -0.0001 | -0.0013 | - | 133.1898 | 111.8596 | |
| ZJZS | 0.0037 | -0.0091 | 0.0037 | - | 16.1058 | |
| GDJS | 0.0100 | 0.0093 | 0.0045 | 0.0301 | - | |
| ND2 | JAP | - | 164.5165 | 57.1701 | 369.8704 | 16.0618 |
| LNDD | 0.0030 | - | ∞ | 34.3918 | 26.5856 | |
| SDQD | 0.0087 | -0.0037 | - | 123.8781 | ∞ | |
| ZJZS | 0.0014 | 0.0143 | 0.0040 | - | 27.6373 | |
| GDJS | 0.0302* | 0.0185 | -0.0103 | 0.0178 | - |
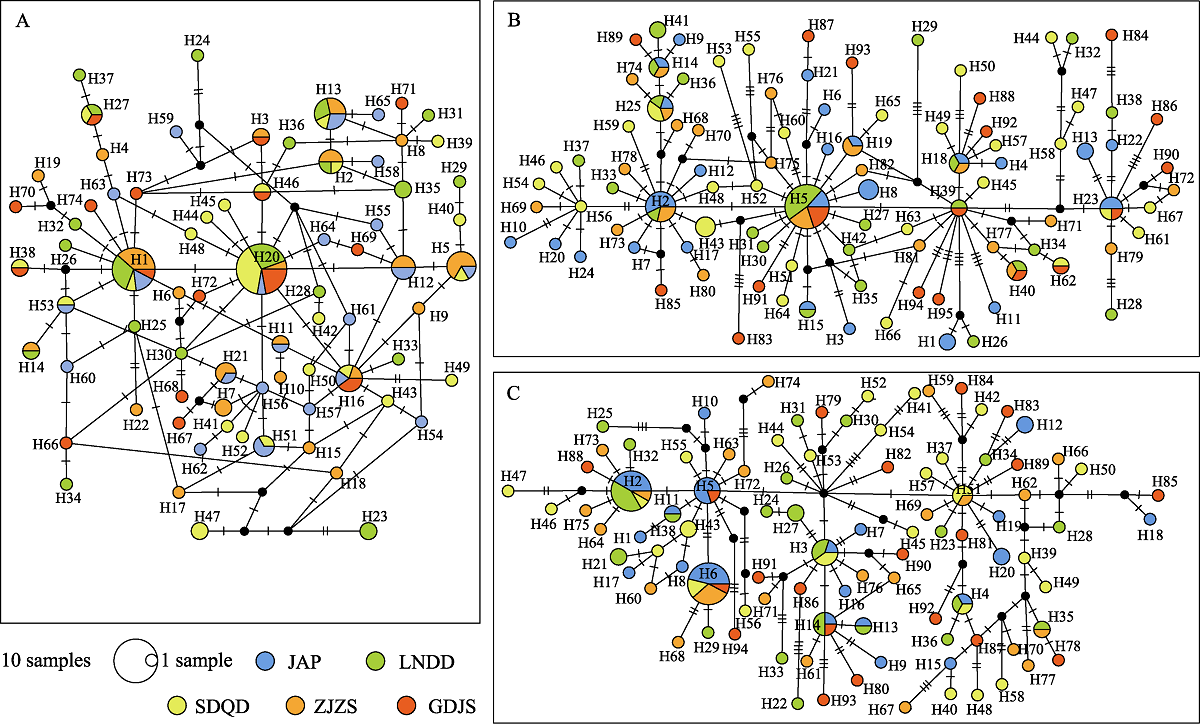
Fig. 2 Median-joining network of mtDNA CR (A), Cytb (B), and ND2 (C) haplotypes from 5 populations of Pleuronichthys cornutus. Different filling traits represent different populations; Node size corresponds to the haplotype frequencies, and “1 sample” size is one individual. Black dots indicate haplotypes that theoretically present but not monitored; Short horizontal line on the line between the circles represents one mutation step. Location codes are the same as the sample sites in Table 1.
| 分组方式 Method of grouping | Cytb/ND2变异比例 Percentage of Cytb/ND2 variation (%) | Cytb/ND2 FST 统计值 Cytb/ND2 FST statistics | ||
|---|---|---|---|---|
| 组群间 Among groups | 组群内群体间 Among populations within groups | 群体内 Within populations | ||
| 7个群体 Seven populations | 0.32/0.85 | -/- | 99.68/99.15 | 0.0032*/0.0085* |
| 中国6个群体和日本1个群体 Six populations from China and one population from Japan | -0.44/0.24 | 0.52/074 | 99.62/99.02 | 0.0008*/0.0098* |
| 濑户内海、中国黄海、东海和南海 Seto inland sea, Yellow Sea, East China Sea, and South China Sea | 0.35/-0.73 | 0/1.50 | 99.64/99.23 | 0.0036*/0.0077* |
| 台湾海峡以南和台湾海峡以北 North of Taiwan Strait and south of Taiwan Strait | 1.87/0.74 | -0.2/0.62 | 98.39/98.64 | 0.0161*/0.0136* |
Table 7 AMOVA analysis of 5 populations from Pleuronichthys cornutus based on Cytb and ND2 sequences. * P < 0.05.
| 分组方式 Method of grouping | Cytb/ND2变异比例 Percentage of Cytb/ND2 variation (%) | Cytb/ND2 FST 统计值 Cytb/ND2 FST statistics | ||
|---|---|---|---|---|
| 组群间 Among groups | 组群内群体间 Among populations within groups | 群体内 Within populations | ||
| 7个群体 Seven populations | 0.32/0.85 | -/- | 99.68/99.15 | 0.0032*/0.0085* |
| 中国6个群体和日本1个群体 Six populations from China and one population from Japan | -0.44/0.24 | 0.52/074 | 99.62/99.02 | 0.0008*/0.0098* |
| 濑户内海、中国黄海、东海和南海 Seto inland sea, Yellow Sea, East China Sea, and South China Sea | 0.35/-0.73 | 0/1.50 | 99.64/99.23 | 0.0036*/0.0077* |
| 台湾海峡以南和台湾海峡以北 North of Taiwan Strait and south of Taiwan Strait | 1.87/0.74 | -0.2/0.62 | 98.39/98.64 | 0.0161*/0.0136* |
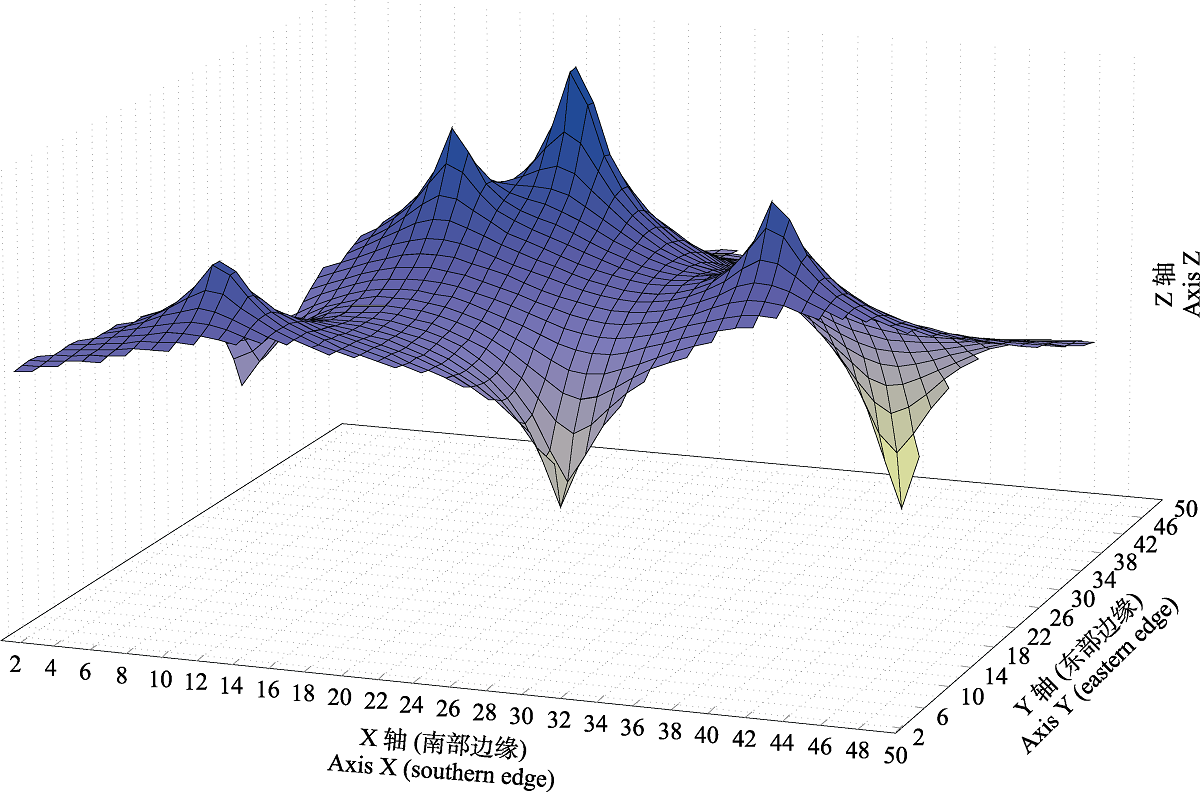
Fig. 3 Genetic landscape shape of Pleuronichthys cornutus based on ND2 gene. X-axis corresponds to longitude and Y-axis corresponds to latitude that for the geographical location of population examined in this study (Table 1); Z-axis corresponds to genetic distance.
| 群体 Population | CR/Cytb/ND2中性检验 CR/Cytb/ND2 neutrality statistics | CR/Cytb/ND2错配分析 CR/Cytb/ND2 mismatch analysis | |||
|---|---|---|---|---|---|
| Tajima’s D | Fu’s FS | SSD | r | τ | |
| JAP | -1.025/-1.751*/-1.624* | -15.980*/-16.752*/-9.805* | 0.011*/0.003/0.003 | 0.044*/0.010/0.019 | 3.3/4.9/5.0 |
| LNDD | -0.961/-1.943*/-1.632* | 13.064*/-15.607*/13.174* | 0.002/0.002/0.014 | 0.017/0.006/0.037 | 3.9/2.4/4.0 |
| SDQD | -0.975/-1.753*/-1.983* | 20.467*/-22.938*/-21.070* | 0.001/0.008/0.006 | 0.019/0.022/0.016 | 3.7/4.0/4.0 |
| ZJZS | -0.105/-1.610*/-1.966* | -21.947*/-19.343*/-17.299* | 0.015*/0.003/0.009 | 0.052*/0.025/0.017 | 3.0/3.1/4.0 |
| GDJS | -1.292/-2.044*/-2.071* | -11.773*/-10.660*/-17.156* | 0.004/0.004/0.016 | 0.032/0.010/0.030 | 3.5/4.0/5.0 |
| 总体 Overall | -0.872/-1.821*/-1.855* | -16.646*/-17.060*/-15.701* | 0.006/0.004/0.010 | 0.033/0.015/0.024 | 3.5/3.7/4.4 |
Table 8 Indices of neutrality statistics and demographic parameter estimates based on CR, Cytb and ND2 sequences in five populations of Pleuronichthys cornutus
| 群体 Population | CR/Cytb/ND2中性检验 CR/Cytb/ND2 neutrality statistics | CR/Cytb/ND2错配分析 CR/Cytb/ND2 mismatch analysis | |||
|---|---|---|---|---|---|
| Tajima’s D | Fu’s FS | SSD | r | τ | |
| JAP | -1.025/-1.751*/-1.624* | -15.980*/-16.752*/-9.805* | 0.011*/0.003/0.003 | 0.044*/0.010/0.019 | 3.3/4.9/5.0 |
| LNDD | -0.961/-1.943*/-1.632* | 13.064*/-15.607*/13.174* | 0.002/0.002/0.014 | 0.017/0.006/0.037 | 3.9/2.4/4.0 |
| SDQD | -0.975/-1.753*/-1.983* | 20.467*/-22.938*/-21.070* | 0.001/0.008/0.006 | 0.019/0.022/0.016 | 3.7/4.0/4.0 |
| ZJZS | -0.105/-1.610*/-1.966* | -21.947*/-19.343*/-17.299* | 0.015*/0.003/0.009 | 0.052*/0.025/0.017 | 3.0/3.1/4.0 |
| GDJS | -1.292/-2.044*/-2.071* | -11.773*/-10.660*/-17.156* | 0.004/0.004/0.016 | 0.032/0.010/0.030 | 3.5/4.0/5.0 |
| 总体 Overall | -0.872/-1.821*/-1.855* | -16.646*/-17.060*/-15.701* | 0.006/0.004/0.010 | 0.033/0.015/0.024 | 3.5/3.7/4.4 |
| [1] |
Ando D, Ikeda M, Sekino M, Sugaya T, Katamachi D, Yoseda K, Kijima A (2016) Improvement of mitochondrial DNA haplotyping in Japanese flounder populations using the sequences of control region and ND2 gene. Nippon Suisan Gakkaishi, 82, 712-719.
DOI URL |
| [2] |
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
PMID |
| [3] | Bao MM, Huo LP, Liu HH (2017) Analysis of the nutritional composition in Pleuronichthys cornutus of Zhoushan. Journal of Anhui Agricultural Sciences, 45(22), 52-54. (in Chinese with English abstract) |
| [包苗苗, 霍利平, 刘慧慧 (2017) 舟山海域角木叶鲽营养成分分析. 安徽农业科学, 45(22), 52-54.] | |
| [4] |
Bradman H, Grewe P, Appleton B (2011) Direct comparison of mitochondrial markers for the analysis of swordfish population structure. Fisheries Research, 109, 95-99.
DOI URL |
| [5] |
Chen DG, Liu CG, Dou SZ (1992) The biology of flatfish (Pleuronectinae) in the coastal waters of China. Netherlands Journal of Sea Research, 29, 25-33.
DOI URL |
| [6] | Dai Q, Lu ZH, Xue LJ, Xu HX (2017) Species composition and assessment of the stock study on Peuronectiformes in the East China Sea. Journal of Zhejiang Ocean University (Natural Science), 36, 379-388. (in Chinese with English abstract) |
| [戴乾, 卢占晖, 薛利建, 徐汉祥 (2017) 东海鲽形目种类组成与资源量评估. 浙江海洋大学学报(自然科学版), 36, 379-388.] | |
| [7] |
Donaldson KA, Wilson RR (1999) Amphi-panamic geminates of snook (Percoidei: Centropomidae) provide a calibration of the divergence rate in the mitochondrial DNA control region of fishes. Molecular Phylogenetics and Evolution, 13, 208-213.
PMID |
| [8] |
Ely B, Viñas J, Bremer J, Black D, Lucas L, Covello K, Labrie A, Thelen E (2005) Consequences of the historical demography on the global population structure of two highly migratory cosmopolitan marine fishes: The yellowfin tuna (Thunnus albacares) and the skipjack tuna (Katsuwonus pelamis). BMC Evolutionary Biology, 5, 19.
DOI URL |
| [9] |
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [10] | Fiedler PL, Jain SK (1992) Conservation Biology: The Theory and Practice of Nature Conservation, Preservation, and Management, 157-158. Chapman & Hall, New York. |
| [11] | Gao TX, Ren GJ, Liu JX, Xiao YS (2009) Progress in molecular phylogeography of marine fish. Periodical of Ocean University of China, 39, 897-902, 1036. (in Chinese with English abstract) |
| [高天翔, 任桂静, 刘进贤, 肖永 (2009) 海洋鱼类分子系统地理学研究进展. 中国海洋大学学报(自然科学版), 39, 897-902, 1036.] | |
| [12] |
Grant W (1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. The Journal of Heredity, 89, 415-426.
DOI URL |
| [13] | Institute of Zoology, Chinese Academy of Sciences, Institute of Oceanology, Chinese Academy of Sciences, Shanghai Ocean University (1962) The Fishes of South China Sea. Science Press, Beijing. (in Chinese) |
| [中国科学院动物研究所, 中国科学院海洋研究所, 上海水产学院 (1962) 南海鱼类志. 科学出版社, 北京.] | |
| [14] | Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution, 1991, 128-144. |
| [15] |
Kocher TD, Conroy JA, Mckaye KR, Stauffer JR, Lockwood S F (1995) Evolution of NADH dehydrogenase subunit 2 in east African cichlid fish. Molecular Phylogenetics and Evolution, 4, 420-432.
PMID |
| [16] | Kong XY, Yu JZ, Zhou LS, Yu ZN (2007) Comparative analysis of 5'-end sequence of the mitochondrial control region of six flatfish species (Pleuronectidae) from the Yellow Sea. Raffles Bulletin of Zoology, 14, 111-120. |
| [17] |
Lee WJ, Conroy J, Howell WH, Kocher TD (1995) Structure and evolution of teleost mitochondrial control regions. Journal of Molecular Evolution, 41, 54-66.
PMID |
| [18] | Li KR (1992) Climate Change and Its Impact in China. China Ocean Press, Beijing. (in Chinese) |
| [李克让 (1992) 中国气候变化及其影响. 海洋出版社, 北京.] | |
| [19] | Li MD (2012) The Ecology of Economic Fishes in China. Tianjin Science and Technology Press, Tianjin. (in Chinese) |
| [李明德 (2012) 中国经济鱼类生态学. 天津科学技术出版社, 天津.] | |
| [20] |
Liu Q, Liu L, Song N, Wang XH, Gao TX (2019) Genetic structure in the marbled rockfish (Sebastiscus marmoratus) across most of the distribution in the northwestern Pacific. Journal of Applied Ichthyology, 35, 1249-1259.
DOI |
| [21] |
Mane S, Guerin P, Mouillot D, Blanchet S, Velez L, Albouy C, Pellissier L (2020) Global determinants of freshwater and marine fish genetic diversity. Nature Communications, 11, 692.
DOI URL |
| [22] |
Miller M (2005) Alleles in space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. The Journal of Heredity, 96, 722-724.
DOI URL |
| [23] |
Ni G, Li Q, Kong LF, Yu H (2014) Comparative phylogeography in marginal seas of the northwestern Pacific. Molecular Ecology, 23, 534-548.
DOI URL |
| [24] |
Okello JBA, Nyakaana S, Masembe C, Siegismund HR, Arctander P (2005) Mitochondrial DNA variation of the common hippopotamus: Evidence for a recent population expansion. Heredity, 95, 206-215.
PMID |
| [25] |
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569.
PMID |
| [26] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution, 34, 3299-3302.
DOI URL |
| [27] |
Song N, Jia N, Yanagimoto T, Lin LS, Gao TX (2013) Genetic differentiation of Trachurus japonicus from the northwestern Pacific based on the mitochondrial DNA control region. Mitochondrial DNA, 24, 705-712.
DOI PMID |
| [28] | Song YX, Gao TX, Yang TY, Han ZQ, Song N (2018) Comparative analysis of genetic diversity and morphology of Pleuronichthys cornutus populations. Periodical of Ocean University of China, 48, 49-55. (in Chinese with English abstract) |
| [宋垚萱, 高天翔, 杨天燕, 韩志强, 宋娜 (2018) 角木叶鲽的群体遗传多样性研究和形态学分析. 中国海洋大学学报(自然科学版), 48, 49-55.] | |
| [29] |
Sudhir K, Glen S, Koichiro T (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874.
DOI PMID |
| [30] | Sun XP (2006) China’s Offshore Area Ocean. China Ocean Press, Beijing. (in Chinese) |
| [孙湘平 (2006) 中国近海区域海洋. 海洋出版社, 北京.] | |
| [31] |
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics, 105, 437-460.
DOI PMID |
| [32] |
Takeshima H, Iguchi K, Nishida M (2005) Unexpected ceiling of genetic differentiation in the control region of the mitochondrial DNA between different subspecies of the Ayu Plecoglossus altivelis. Zoological science, 22, 401-410.
PMID |
| [33] |
Tarallo A, Angelini C, Sanges R, Yagi M, Agnisola C, D’Onofrio G (2016) On the genome base composition of teleosts: The effect of environment and lifestyle. BMC Genomics, 17, 173.
DOI PMID |
| [34] | Verma R, Singh M, Kumar S (2016) Unraveling the limits of mitochondrial control region to estimate the fine scale population genetic differentiation in anadromous fish Tenualosa ilisha. Scientifica, 2016, 2035240. |
| [35] |
Wang L, Shi XF, Su YQ, Meng ZN, Lin HR (2013) Genetic divergence and historical demography in the endangered large yellow croaker revealed by mtDNA. Biochemical Systematics and Ecology, 46, 137-144.
DOI URL |
| [36] |
Wang PX (1999) Response of western Pacific marginal seas to glacial cycles: Paleoceanographic and sedimentological features. Marine Geology, 156, 5-39.
DOI URL |
| [37] | Wright S (1978) Evolution and the Genetics of Population:Variability Within and Among Natural Population. University of Chicago Press, Chicago. |
| [38] |
Xiang DG, Li YF, Li XH, Chen WT, Ma XH (2021) Population structure and genetic diversity of Culter recurviceps revealed by multi-loci. Biodiversity Science, 29, 1505-1512. (in Chinese with English abstract)
DOI URL |
|
[向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧 (2021) 多基因联合揭示海南鲌的遗传结构与遗传多样性. 生物多样性, 29, 1505-1512.]
DOI |
|
| [39] | Xiao WH, Zhang YP (2000) Genetics and evolution of mitochondrial DNA in fish. Acta Hydrobiologica Sinica, 24, 385- 391. (in Chinese with English abstract) |
| [肖武汉, 张亚平 (2000) 鱼类线粒体DNA的遗传与进化. 水生生物学报, 24, 385-391.] | |
| [40] |
Yamamoto K, Nagasawa K (2015) Temporal changes in demersal fish assemblage structure in the East China Sea and the Yellow Sea. Nippon Suisan Gakkaishi, 81, 429-437.
DOI URL |
| [41] | Yang XS, Zhang Q, Yu FY, Lü JL, Di XD, Shao WJ, Huang ZY, Lu LF (2017) MtDNA ND2 sequence-based genetic analysis of Anabas testudineus from South China and Lancang Mekong River. South China Fisheries Science, 13, 43-50. (in Chinese with English abstract) |
| [杨喜书, 章群, 余帆洋, 吕金磊, 底晓丹, 邵伟军, 黄镇宇, 卢丽锋 (2017) 华南6水系与澜沧江-湄公河攀鲈线粒体ND2基因的遗传多样性分析. 南方水产科学, 13, 43-50.] | |
| [42] | Yao YT, Harff J, Meyer M, Zhan WH (2009) Reconstruction of paleocoastlines for the northwestern South China Sea since the Last Glacial Maximum. Science in China Series D: Earth Sciences, 39, 753-762. (in Chinese with English abstract) |
| [姚衍桃, Harff Jan, 詹文欢 (2009) 南海西北部末次盛冰期以来的古海岸线重建. 中国科学(D辑: 地球科学), 39, 753-762.] | |
| [43] | Yue XL, Zhang Q, Zhao S, Fan FJ (2010) A fast and efficient method for isolation of genomic DNA from fish specimens. Biotechnology Bulletin, (2), 202-204. (in Chinese with English abstract) |
| [乐小亮, 章群, 赵爽, 范凤娟 (2010) 一种高效快速的鱼类标本基因组DNA提取方法. 生物技术通报, (2), 202-204.] | |
| [44] |
Zardoya R, Meyer A (1996) Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Molecular Biology and Evolution, 13, 933-942.
PMID |
| [45] | Zheng DY, Guo YJ, Yang TY, Gao TX, Zheng Y, Yuan DH, Si SJ (2019) Genetic diversity analysis of Sillago japonica based on mitochondrial DNA ND2 gene. South China Fisheries Science, 15, 84-91. (in Chinese with English abstract) |
| [郑德育, 郭易佳, 杨天燕, 高天翔, 郑瑶, 袁冬皓, 斯舒谨 (2019) 基于线粒体ND2基因序列的少鳞鱚遗传多样性研究. 南方水产科学, 15, 84-91.] | |
| [46] | Zhu Y, Zhang Q, Li GS, Liu HL, Ma B, Huang XY, Si CL (2012) Genetic diversity of four Pleuronichthys cornutus populations in coastal waters of China. Marine Science Bulletin, 31, 552-556. (in Chinese with English abstract) |
| [朱叶, 章群, 李贵生, 刘海林, 马奔, 黄小彧, 司从利 (2012) 中国近海角木叶鲽(Pleuronichthys cornutus)种群遗传多样性研究. 海洋通报, 31, 552-556.] | |
| [47] | Zhu YD, Zhang CL (1963) The Fishes of East China Sea. Science Press, Beijing. (in Chinese) |
| [朱元鼎, 张春霖 (1963) 东海鱼类志. 科学出版社, 北京.] |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [11] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [12] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [13] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [14] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| [15] | Jun Sun, Yuyao Song, Yifeng Shi, Jian Zhai, Wenzhuo Yan. Progress of marine biodiversity studies in China seas in the past decade [J]. Biodiv Sci, 2022, 30(10): 22526-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()