



生物多样性 ›› 2015, Vol. 23 ›› Issue (6): 784-792. DOI: 10.17520/biods.2015075 cstr: 32101.14.biods.2015075
所属专题: 青藏高原生物多样性与生态安全
谢理丽, 徐磊*, 林秋奇, 韩博平
收稿日期:2015-03-25
接受日期:2015-09-01
出版日期:2015-11-20
发布日期:2015-12-02
通讯作者:
徐磊
基金资助:Lili Xie, Lei Xu*, Qiuqi Lin, Boping Han
Received:2015-03-25
Accepted:2015-09-01
Online:2015-11-20
Published:2015-12-02
Contact:
Xu Lei
摘要:
本文通过对西藏湖泊长刺溞复合种(Daphnia longispina complex)中分布最广的3个物种, 即长刺溞(D. longispina)、盔形溞(D. galeata)和颈齿溞(D. dentifera)线粒体COI基因序列以及GenBank中欧洲的长刺溞、加拿大的颈齿溞和我国东部低海拔地区的盔型溞COI基因序列的比较分析, 研究了西藏湖泊长刺溞复合种的系统进化关系, 发现西藏地区的盔型溞、颈齿溞和长刺溞均已出现较大分化。颈齿溞种群内遗传差异度为0.33-2.32%, 盔型溞为0.33-2.74%, 长刺溞的遗传差异度最高, 为1.31-5.50%。基于COI基因序列构建的最大似然树和贝叶斯系统树均表明, 长刺溞复合种由3个进化分支组成, 分别对应长刺溞、盔型溞和颈齿溞, 三者之间的遗传差异度为9.40-16.98%(Kimura 2-parameter双参数模型)。基于COI基因单倍型(haplotype)所构建的网络关系也支持上述3个分支的存在。早期记录虽然显示长刺溞在我国分布较广, 但本次调查只在班公错有发现, 相比之下, 盔形溞和颈齿溞则分布更广。我们的研究表明, 由于形态学鉴定上的局限性, 早期的长刺溞记录很可能混杂了容易引起混淆的盔型溞或颈齿溞。
谢理丽, 徐磊, 林秋奇, 韩博平 (2015) 西藏湖泊长刺溞复合种的系统进化关系. 生物多样性, 23, 784-792. DOI: 10.17520/biods.2015075.
Lili Xie, Lei Xu, Qiuqi Lin, Boping Han (2015) Phylogenetics of the Daphnia longispina complex in Tibetan lakes. Biodiversity Science, 23, 784-792. DOI: 10.17520/biods.2015075.
| 采样点 Location | 纬度 经度 Latitude Longitude | 代号 Code | GenBank登录号 GenBank accession | 序列来源 Sequence source |
|---|---|---|---|---|
| 盔形溞 Daphnia galeata | ||||
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE1 | 本研究 This study | |
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE2 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC1 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC2 | 本研究 This study | |
| 骆马湖 Luoma Lake, China | 34.07º N 118.11° E | HC3 | KM555356 | GenBank |
| 西湖 West Lake, China | 30.15º N 120.08° E | HC2 | KM555355 | GenBank |
| 宝应湖 Baoying Lake, China | 33.10º N 119.14° E | HC1 | KM555354 | GenBank |
| 颈齿溞 Daphnia dentifera | ||||
| 沉错 Chencuo, China | 28.88º N 90.47° E | CC | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS1 | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS2 | 本研究 This study | |
| 那曲湖 Naqu Lake, China | 31.48º N 92.05° E | NQHA | KM555369 | GenBank |
| Canada | CND | FJ427488 | GenBank | |
| 格桑桥 Gesangqiao, China | 29.65º N 91.12° E | GSQA | KM555366 | GenBank |
| 长刺溞 Daphnia longispina | ||||
| 班公错 Bangongcuo, China 班公错 Bangongcuo, China | 33.71º N 78.81° E 33.71º N 78.81° E | BGC1 BGC2 | 本研究 This study 本研究 This study | |
| Sweden Germany Switzerland | DEM H29 SZL | EF375861 EF375860 EF375862 | GenBank GenBank GenBank |
表1 长刺溞复合种种群位点及序列信息
Table 1 Geographic and genetic characteristics of 18 Daphnia longispina complex populations
| 采样点 Location | 纬度 经度 Latitude Longitude | 代号 Code | GenBank登录号 GenBank accession | 序列来源 Sequence source |
|---|---|---|---|---|
| 盔形溞 Daphnia galeata | ||||
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE1 | 本研究 This study | |
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE2 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC1 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC2 | 本研究 This study | |
| 骆马湖 Luoma Lake, China | 34.07º N 118.11° E | HC3 | KM555356 | GenBank |
| 西湖 West Lake, China | 30.15º N 120.08° E | HC2 | KM555355 | GenBank |
| 宝应湖 Baoying Lake, China | 33.10º N 119.14° E | HC1 | KM555354 | GenBank |
| 颈齿溞 Daphnia dentifera | ||||
| 沉错 Chencuo, China | 28.88º N 90.47° E | CC | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS1 | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS2 | 本研究 This study | |
| 那曲湖 Naqu Lake, China | 31.48º N 92.05° E | NQHA | KM555369 | GenBank |
| Canada | CND | FJ427488 | GenBank | |
| 格桑桥 Gesangqiao, China | 29.65º N 91.12° E | GSQA | KM555366 | GenBank |
| 长刺溞 Daphnia longispina | ||||
| 班公错 Bangongcuo, China 班公错 Bangongcuo, China | 33.71º N 78.81° E 33.71º N 78.81° E | BGC1 BGC2 | 本研究 This study 本研究 This study | |
| Sweden Germany Switzerland | DEM H29 SZL | EF375861 EF375860 EF375862 | GenBank GenBank GenBank |
| HC3 | HC1 | HC2 | CE | DJMC | |
|---|---|---|---|---|---|
| HC3 | n.c. | ||||
| HC1 | 0.33 ± 0.23 | n.c. | |||
| HC2 | 2.15 ± 0.57 | 1.81 ± 0.53 | n.c. | ||
| CE | 2.74 ± 0.66 | 2.40 ± 0.63 | 0.90 ± 0.36 | 0.16 ± 0.15 | |
| DJMC | 2.49 ± 0.62 | 2.15 ± 0.59 | 0.65 ± 0.33 | 0.90 ± 0.37 | 0.16 ±0.15 |
表2 盔形溞(Daphnia galeata)种群间(对角线下)和种群内(对角线)的遗传差异度(%)
Table 2 The genetic differentiation (%) between (below the diagonal) and within (on the diagonal) Daphnia galeata populations
| HC3 | HC1 | HC2 | CE | DJMC | |
|---|---|---|---|---|---|
| HC3 | n.c. | ||||
| HC1 | 0.33 ± 0.23 | n.c. | |||
| HC2 | 2.15 ± 0.57 | 1.81 ± 0.53 | n.c. | ||
| CE | 2.74 ± 0.66 | 2.40 ± 0.63 | 0.90 ± 0.36 | 0.16 ± 0.15 | |
| DJMC | 2.49 ± 0.62 | 2.15 ± 0.59 | 0.65 ± 0.33 | 0.90 ± 0.37 | 0.16 ±0.15 |
| DEM | H29 | SZL | BGC | |
|---|---|---|---|---|
| DEM | n.c. | |||
| H29 | 1.81 ± 0.53 | n.c. | ||
| SZL | 1.81 ± 0.54 | 1.31 ± 0.43 | n.c. | |
| BGC | 5.50 ± 0.92 | 4.88 ± 0.84 | 4.62 ± 0.80 | 1.62 ± 0.51 |
表3 长刺溞(Daphnia longispina)种群间(对角线下)和种群内(对角线)的遗传差异度(%)
Table 3 The genetic differentiation (%) between (below the diagonal) and within (on the diagonal) Daphnia longispina populations
| DEM | H29 | SZL | BGC | |
|---|---|---|---|---|
| DEM | n.c. | |||
| H29 | 1.81 ± 0.53 | n.c. | ||
| SZL | 1.81 ± 0.54 | 1.31 ± 0.43 | n.c. | |
| BGC | 5.50 ± 0.92 | 4.88 ± 0.84 | 4.62 ± 0.80 | 1.62 ± 0.51 |
| CND | NQHA | GSQA | LS | CC | |
|---|---|---|---|---|---|
| CND | n.c. | ||||
| NQHA | 1.98 ± 0.55 | n.c. | |||
| GSQA | 2.15 ± 0.60 | 0.49 ± 0.30 | n.c. | ||
| LS | 2.32 ± 0.60 | 0.65 ± 0.30 | 0.33 ± 0.16 | 0.65 ± 0.32 | |
| CC | 2.15 ± 0.60 | 0.49 ± 0.30 | 0.00 | 0.33 ± 0.16 | n.c. |
表4 颈齿溞(Daphnia dentifera)种群间(对角线下)和种群内(对角线)的遗传差异度(%)
Table 4 The genetic differentiation (%) between (below the diagonal) and within (on the diagonal) Daphnia dentifera populations
| CND | NQHA | GSQA | LS | CC | |
|---|---|---|---|---|---|
| CND | n.c. | ||||
| NQHA | 1.98 ± 0.55 | n.c. | |||
| GSQA | 2.15 ± 0.60 | 0.49 ± 0.30 | n.c. | ||
| LS | 2.32 ± 0.60 | 0.65 ± 0.30 | 0.33 ± 0.16 | 0.65 ± 0.32 | |
| CC | 2.15 ± 0.60 | 0.49 ± 0.30 | 0.00 | 0.33 ± 0.16 | n.c. |
| DG | DL | DD | |
|---|---|---|---|
| DG | 1.46 ± 0.31 | 71(67.62%) | 79(80.61%) |
| DL | 16.70 ± 1.72 | 3.66 ± 0.59 | 32(49.23%) |
| DD | 16.98 ± 1.87 | 9.40 ± 1.12 | 1.01 ± 0.24 |
表5 长刺溞复合种(Daphnia longispina complex)种间和种内的遗传差异度(%) (对角线和对角线下)及复合种间各单倍型的固化差异(对角线上)
Table 5 The genetic differentiation (%) between and within species in Daphnia longispina complex (on the diagonal and below the diagonal) and the fixed differences among these haplotypes (above the diagonal)
| DG | DL | DD | |
|---|---|---|---|
| DG | 1.46 ± 0.31 | 71(67.62%) | 79(80.61%) |
| DL | 16.70 ± 1.72 | 3.66 ± 0.59 | 32(49.23%) |
| DD | 16.98 ± 1.87 | 9.40 ± 1.12 | 1.01 ± 0.24 |
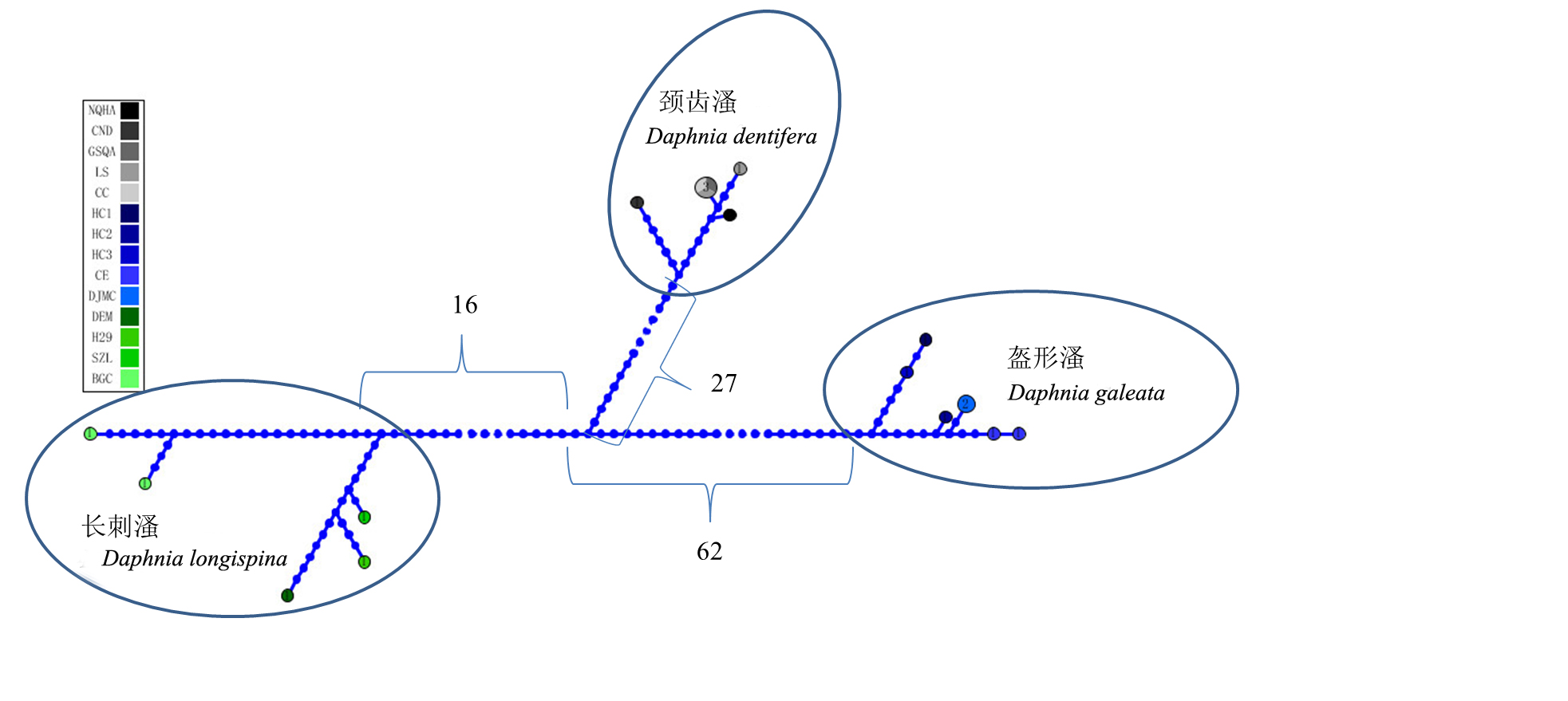
图1 基于COI基因序列构建的长刺溞复合种单倍型网络图(图中数字代表了各分支的平均突变数量)。图例中的缩写参见表1。
Fig. 1 Haplotype network for Daphnia longispina complex based on mitochondria COI gene sequences. Numbers between subnetworks represent the average number of mutations between subclades. The population codes are the same as those in Table 1.
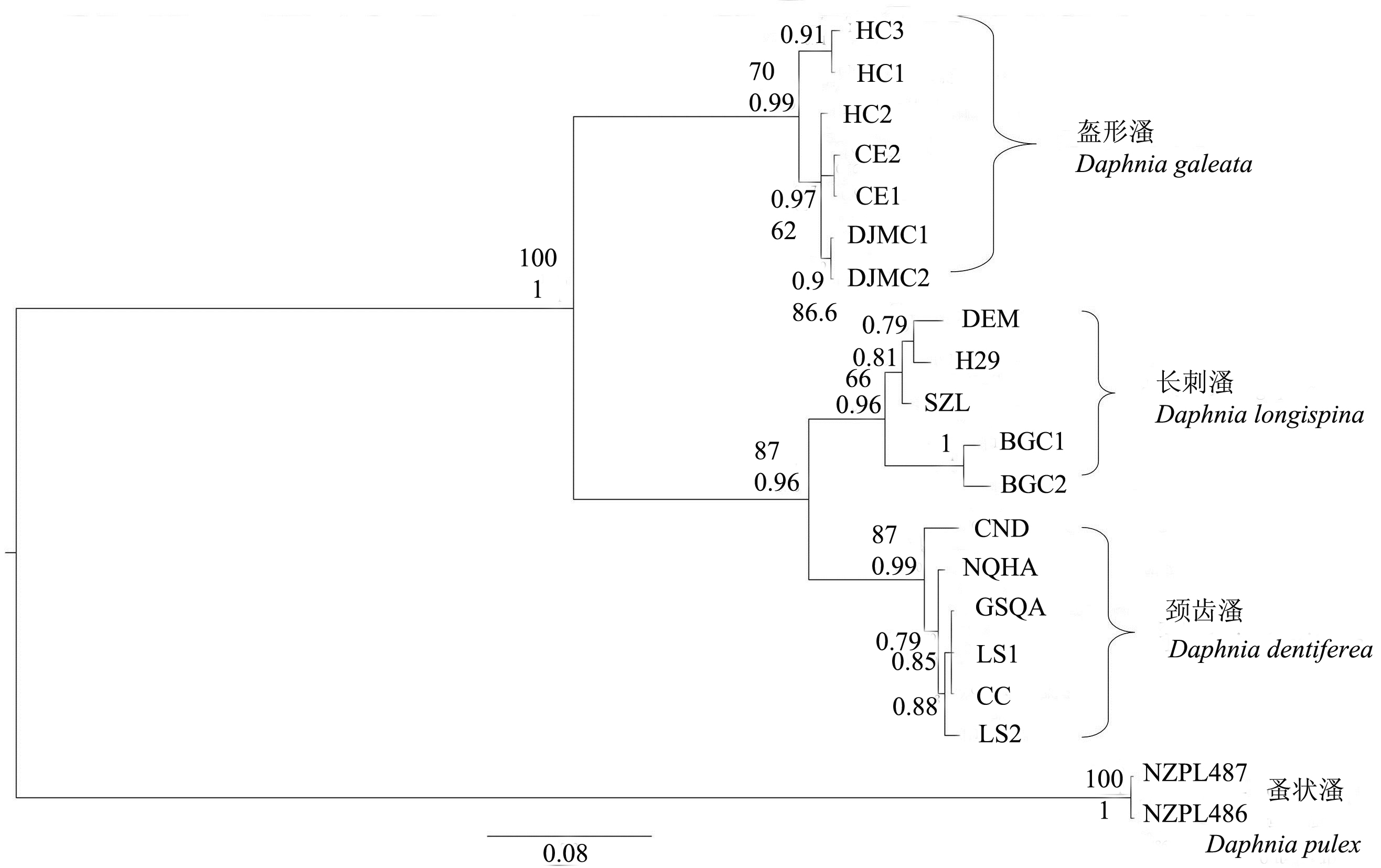
图2 基于COI基因序列构建长刺溞复合种贝叶斯系统树与最大似然法系统树组合的进化树, 贝叶斯系统树后验概率(PP)和最大似然法系统进化树的自举支持率(bootstrap values)(百分数)均标于树分支的上面。图中的缩写为表1中COI序列代号。
Fig. 2 Bayes and Maximum Likelihood phylogenetic tree of Daphnia longispina complex based on mitochondria COI gene sequences. Posterior probabilities (Bayes tree) and bootstrap values (Maximum Likelihood tree) are shown above the nodes. The population codes are the same as those in Table 1.
| 1 | Adamowicz SJ, Purvis A (2005) How many branchiopod crustacean species are there? Quantifying the components of underestimation.Global Ecology and Biogeography, 14, 455-468. |
| 2 | Adamowicz SJ, Petrusek A, Colbourne JK, Hebert PDN, Witt JDS (2009) The scale of divergence: a phylogenetic appraisal of intercontinental allopatric speciation in a passively dispersed freshwater zooplankton genus.Molecular Phylogenetics and Evolution, 50, 423-436. |
| 3 | Allen MR, Thum RA, Caceres CE (2010) Does local adaptation to resources explain genetic differentiation among Daphnia populations?Molecular Ecology, 19, 3076-3087. |
| 4 | Benzie JAH (1988) The systematics of Australian Daphnia (Cladocera: Daphniidae). Electrophoretic analyses of the Daphnia carinata complex.Hydrobiologia, 166, 183-197. |
| 5 | Benzie JAH (2005) Cladocera: the genus Daphnia (including Daphniopsis) (Anomopoda: Daphniidae). Guides to the identification of the microinvertebrates of the continental waters of the world.Quarterly Review of Biology, 80, 491-510. |
| 6 | Chiang SC (蒋燮治), Du NS (堵南山) (1979) Fauna Sinica, Crustacea, Freshwater Cladocera (中国动物志·淡水枝角类). Science Press, Beijing. (in Chinese) |
| 7 | Chiang SC (蒋燮治), Shen YF (沈韫芬), Gong XJ (龚循矩) (1983) Aquatic Invertebrates of the Tibetan Plateau (西藏水生无脊椎动物). Science Press, Beijing. (in Chinese) |
| 8 | Colbourne JK, Crease TJ, Weider LJ, Hebert PDN, Dufresne F, Hobaek A (1998) Phylogenetics and evolution of a circumarctic species complex (Cladocera: Daphnia pulex).Biological Journal of the Linnean Society, 65, 347-365. |
| 9 | Costa FO, Jeremy RD, James B, Rantnasingham S, Dooh RT, Hajibaei M, Hebert PDN (2007) Biological identifications through DNA barcodes: the case of the Crustacea.Canadian Journal of Fisheries and Aquatic Sciences, 64, 272-290. |
| 10 | De Meester L (1996) Local genetic differentiation and adaptation in freshwater zooplankton populations: patterns and processes.Ecoscience, 3, 385-399. |
| 11 | Dlouhá S, Thielsch A, Kraus RHS, Seda J, Schwenk K, Petrusek A (2010) Identifying hybridizing taxa within the Daphnia longispina species complex: which methods to rely on?Hydrobiologia, 643, 107-122. |
| 12 | Dodson S (1990) Predicting diel vertical migration of zooplankton.Limnology and Oceanography, 35, 1195-1200. |
| 13 | Dumont HJ, Negrea S (2002) Introduction to the Class Brachiopoda, Guides to the Identification of the Microinvertebrates of the Continental Waters of World 19. Backhuys Publishers, Leiden. |
| 14 | Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates.Molecular Marine Biology and Biotechnology, 3, 294-299. |
| 15 | Forro′ L, Korovchinsky NM, Kotov AA, Petrusek A (2008) Global diversity of cladocerans (Cladocera; Crustacea) in freshwater. Hydrobiologia, 595, 177-184. |
| 16 | Giessler S (1997a) Analysis of reticulate relationships within the Daphnia longispina species complex. Allozyme phenotype and morphology.Journal of Evolutionary Biology, 10, 87-105. |
| 17 | Giessler S (1997b) Gene flow in the Daphnia longispina hybrid complex (Crustacea, Cladocera) inhabiting large lakes.Heredity, 79, 231-241. |
| 18 | Hebert PHD (1978) The population biology of Daphnia (Crustacea, Daphnidae).Biological Reviews, 53, 387-426. |
| 19 | Hebert PDN, Cywinska A, Ball SL, De Waard JR (2003a) Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. |
| 20 | Hebert PDN, Witt JDS, Adamowicz SJ (2003b) Phylogeographical patterning in Daphnia ambigua: regional divergence and intercontinental cohesion.Limnology and Oceanography, 48, 261-268. |
| 21 | Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny.Bioinformatics, 17, 754-755. |
| 22 | Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science, 294, 2310-2314. |
| 23 | Hutchison GE (1967) A Treatise on Limnology. PhD dissertation, National Academy of Sciences, New York. |
| 24 | Irestedt M, Fjeldså J, Nylander J, Ericson P (2004) Phylogenetic relationships of typical antbirds (Thamnophilidae) and test of incongruence based on Bayes factors.BMC Evolutionary Biology, 4, 1-16. |
| 25 | Ishida S, Taylor DJ (2007) Quaternary diversification in a sexual Holarctic zooplankter, Daphnia galeata.Molecular Ecology, 16, 569-582. |
| 26 | Ishida S, Takahashi A, Matsushima N, Yokoyama J, Makino W, Urabe J, Kawata M (2011) The long-term consequences of hybridization between the two Daphnia species, D. galeata and D. dentifera, in mature habitats.BMC Evolutionary Biology, 11, 209-222. |
| 27 | Korovchinsky NM (1996) How many species of Cladocera are there?Hydrobiologia, 321, 191-204. |
| 28 | Kotov AA (2015) A critical review of the current taxonomy of the genus Daphnia O. F. Müller, 1785 (Anomopoda, Cladocera).Zootaxa, 3911, 184-200. |
| 29 | Kotov AA, Ishida S, Taylor DJ (2006) A new species in the Daphnia curvirostris (Crustacea: Cladocera) complex from the eastern Palearctic with molecular phylogenetic evidence for the independent origin of neckteeth.Journal of Plankton Research, 28, 1067-1079. |
| 30 | Kumar S, Dudley J, Nei M, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequence.Briefings in Bioinformatics, 9, 299-306. |
| 31 | Lampert W (2011) Daphnia: Development of Model Organism in Ecology and Evolution. American Institute of Biological Sciences, Washington DC. |
| 32 | Ma XL, Petrusek A, Wolinska J, Gießler S, Zhong Y, Hu W, Yin MB (2015) Diversity of the Daphnia longispina species complex in Chinese lakes: a DNA taxonomy approach.Journal of Plankton Research, 37, 56-65. |
| 33 | Mergeay J, Aguilera X, Declerck S, Petrusek A, Huyse T, De Meester L (2008) The genetic legacy of polyploid Bolivian Daphnia: the tropical Andes as a source for the North and South American Daphnia pulicaria complex.Molecular Ecology, 17, 1789-1800. |
| 34 | Mőst M, Petrusek A, Sommaruga R, Juračka PJ, Slusarczyk M, Manca M (2013) At the edge and on the top: molecular identification and ecology of Daphnia dentifera and D. longispina in high-altitude Asian lakes.Hydrobiologia, 715, 169-180. |
| 35 | Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers.Computer Applications in the Biosciences, 12, 357-358. |
| 36 | Petrusek A, Cerny M, Mergeay J, Schwenk K (2007) Daphnia in the Tatra Mountain lakes: multiple colonization and hidden species diversity revealed by molecular markers.Fundamental and Applied Limnology, 169, 279-291. |
| 37 | Petrusek A, Hobæk A, Nilssen JP, Skage M, Černý M, Brede N, Schwenk K (2008) A taxonomic reappraisal of the European Daphnia longispina complex (Crustacea, Cladocera, Anomopoda).Zoologica Scripta, 37, 507-519. |
| 38 | Petrusek A (2007) Diversity of European Daphnia on Different Scales: from Cryptic Species to Within-lake Differentiation. PhD dissertation, Charles University, Prague. |
| 39 | Posada D, Crandall KA (2001) Selecting the best-fit model of nucleotide substitution.Systematic Biology, 50, 580-601. |
| 40 | Rozas J, Sanchez-Delbarria JC, Messeguer X (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods.Bioinformatics, 19, 2496-2497. |
| 41 | Salzburger W, Ewing GB, Haeseler VA (2011) The performance of phylogenetic algorithms in estimating haplotype genealogies with migration.Molecular Ecology, 20, 1952-1963. |
| 42 | Sekino T, Yoshioka T (1995) The relationship between nutritional condition and diel vertical migration of Daphnia galeata.Japanese Journal of Limnology, 56, 145-150. |
| 43 | Skage M, Hobæk A, Ruthová Š, Keller B, Petrusek A, Sed’a J, Spaak P (2007) Intra-specific rDNA-ITS restriction site variation and an improved protocol to distinguish species and hybrids in the Daphnia longispina complex.Hydrobiologia, 594, 19-32. |
| 44 | Sommer RS, Zachos FE (2009) Fossil evidence and phylogeography of temperate species: glacial refugia and post-glacial recolonization.Journal of Biogeography, 36, 2013-2020. |
| 45 | Spaak P, Fox J, Hairston NG Jr (2012) Modes and mechanisms of a Daphnia invasion.Proceedings of the Royal Society B: Biological Sciences, 279, 2936-2944. |
| 46 | Swofford D (2003) PAUP* 4.0 |
| [Electronic Resource]: Phylogenetic Analysis Using Parsimony. Sinauer Associates Publishers, Sunderland. | |
| 47 | Taylor DJ, Hebert PD, Colbourne JK (1996) Phylogenetics and evolution of the Daphnia longispina group (Crustacea) based on 12S rDNA sequence and allozyme variation.Molecular Phylogenetics and Evolution, 5, 495-510. |
| 48 | Taylor DJ, Finston TL, Hebert PDN (1998) Biogeography of a widespread freshwater crustacean: pseudocongruence and cryptic endemism in the North American Daphnia laevis complex.Evolution, 52, 1648-1670. |
| 49 | Ventura M, Petrusek A, Miró A, Hamrová E, Buñay D, De Meester L, Mergeay J (2014) Local and regional founder effects in lake zooplankton persist after thousands of years despite high dispersal potential.Molecular Ecology, 23, 1014-1027. |
| 50 | Xu L (徐磊), Li SJ (李思嘉), Wang S (王晟), Han XY (韩小玉), Han BP (韩博平) (2014) Comparison of two methods for extracting DNA of resting eggs by Cladocera.Journal of Lake Sciences(湖泊科学), 26, 632-636. (in Chinese with English abstract) |
| 51 | Xu L (徐磊) (2013) Biogeography and Genetic Diversity of Two Cladoceran Species (Leptodora kindtii and Daphnia galeata) (两种枝角类(Leptodora kindtii 和 Daphnia galeata)的生物地理学及其种群遗传多样性研究). PhD dissertation, Jinan University, Guangzhou. (in Chinese with English abstract) |
| 52 | Zhang JC (张继承), Jiang QG (姜琦刚), Li YH (李远华), Wang K (王坤) (2008) Dynamic monitoring and climatic background of lake changes in Tibet based on RS/GIS.Journal of Earth Sciences and Environment(地球科学与环境学报), 30, 285-292. (in Chinese with English abstract) |
| [1] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [2] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [3] | 陈又生, 宋柱秋, 卫然, 罗艳, 陈文俐, 杨福生, 高连明, 徐源, 张卓欣, 付鹏程, 向春雷, 王焕冲, 郝加琛, 孟世勇, 吴磊, 李波, 于胜祥, 张树仁, 何理, 郭信强, 王文广, 童毅华, 高乞, 费文群, 曾佑派, 白琳, 金梓超, 钟星杰, 张步云, 杜思怡. 西藏维管植物多样性编目和分布数据集[J]. 生物多样性, 2023, 31(9): 23188-. |
| [4] | 卢聪聪, 刘倩, 黄晓磊. 三种蚜虫的线粒体基因组数据[J]. 生物多样性, 2022, 30(7): 22204-. |
| [5] | 翟俊杰, 赵慧峰, 商光申, 孙振钧, 张玉峰, 王兴. 蚯蚓基因组学的研究进展: 基于全基因组及线粒体基因组[J]. 生物多样性, 2022, 30(12): 22257-. |
| [6] | 张爱梅, 殷一然, 孔维宝, 朱学泰, 杨颖丽. 西藏沙棘5种不同组织内生细菌多样性[J]. 生物多样性, 2021, 29(9): 1236-1244. |
| [7] | 李潮,金锦锦,罗锦桢,王春晖,王俊杰,赵俊. 唐鱼养殖种群与广州附近4个野生种群的遗传关系[J]. 生物多样性, 2020, 28(4): 474-484. |
| [8] | 刘成, 亚吉东, 郭永杰, 蔡杰, 张挺. 西藏种子植物分布新资料[J]. 生物多样性, 2020, 28(10): 1238-1245. |
| [9] | 王渊, 李晟, 刘务林, 朱雪林, 李炳章. 西藏雅鲁藏布大峡谷国家级自然保护区金猫的色型类别与活动节律[J]. 生物多样性, 2019, 27(6): 638-647. |
| [10] | 杨计平, 李策, 陈蔚涛, 李跃飞, 李新辉. 西江中下游鳤的遗传多样性与种群动态历史[J]. 生物多样性, 2018, 26(12): 1289-1295. |
| [11] | 于少帅, 徐启聪, 林彩丽, 王圣洁, 田国忠. 植原体遗传多样性研究现状与展望[J]. 生物多样性, 2016, 24(2): 205-215. |
| [12] | 徐晓婷, 王志恒, DimitarDimitrov. 批量下载GenBank基因序列数据的新工具——NCBIminer[J]. 生物多样性, 2015, 23(4): 550-555. |
| [13] | 牛红玉, 王峥峰, 练琚愉, 叶万辉, 沈浩. 群落构建研究的新进展: 进化和生态 相结合的群落谱系结构研究[J]. 生物多样性, 2011, 19(3): 275-283. |
| [14] | 杨军, Humphrey G. Smith, David M. Wilkinson. 北极、南极和西藏有壳虫区系与分布[J]. 生物多样性, 2010, 18(4): 373-382. |
| [15] | 赵阿曼, 刘志民, 康向阳, 周世良. 西藏特有植物砂生槐天然居群遗传多样性研究[J]. 生物多样性, 2003, 11(2): 91-99. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2026 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()