



生物多样性 ›› 2025, Vol. 33 ›› Issue (8): 25184. DOI: 10.17520/biods.2025184 cstr: 32101.14.biods.2025184
收稿日期:2025-05-19
接受日期:2025-08-28
出版日期:2025-08-20
发布日期:2025-09-17
通讯作者:
*E-mail: huangxl@fafu.edu.cn
基金资助:
Xin Peng( ), Chuan Liu(
), Chuan Liu( ), Xiaolei Huang*(
), Xiaolei Huang*( )(
)( )
)
Received:2025-05-19
Accepted:2025-08-28
Online:2025-08-20
Published:2025-09-17
Contact:
*E-mail: huangxl@fafu.edu.cn
Supported by:摘要:
遗传数据在生物多样性研究和保护实践中发挥着越来越重要的作用, 然而研究者应用这些数据时常面临数据质量缺陷、地理或类群分布不均等方面的制约, 尽管陆生脊椎动物的遗传数据格局已有较深入研究, 但全球植物、真菌和其他动物类群的遗传数据空间分布模式仍缺乏系统的实证研究。本文采用多尺度分析方法, 系统评估了动物、植物和真菌三大真核生物界的遗传数据现状、元数据完整性以及遗传数据的时空动态趋势。结果表明, 动物界拥有约2.7亿条序列和1.6万个基因组数据, 超过植物界(约1.4亿条, 0.7万个)和真菌界(约0.2亿条, 1.7万个)。遗传数据的地理元数据缺失现象普遍存在, 其中真菌ITS序列的经纬度缺失最为严重(缺失率92.07%), 其次是植物rbcL (83.19%)和动物COI (26.40%)。时空分布格局显示, 全球尺度上遗传数据呈现明显的“北半球中心化”特征, 北美、西欧和东亚地区占据主导地位, 而南半球普遍数据匮乏; 同时观察到动物COI和植物rbcL数据呈下降趋势, 而真菌ITS数据快速增长。中国区域则表现出独特的“南动物、东植物、北真菌”分布格局, 而西北地区数据积累明显不足; 时间维度上, 中国植物和真菌数据持续增长, 而动物数据保持稳定。这些发现揭示了遗传数据质量缺陷和分布失衡已成为制约生物多样性研究的重要瓶颈。为此, 我们建议建立严格的元数据存档标准, 重点加强南半球和中国西北部等数据薄弱区域的科研投入, 并通过构建国际科研合作网络促进全球数据资源的均衡配置, 从而提升遗传数据在生物多样性研究和保护实践中的应用价值。
彭欣, 刘传, 黄晓磊 (2025) 基于GenBank数据库的真核生物遗传数据时空格局分析. 生物多样性, 33, 25184. DOI: 10.17520/biods.2025184.
Xin Peng, Chuan Liu, Xiaolei Huang (2025) Spatiotemporal pattern analysis of eukaryotic genetic data based on the GenBank database. Biodiversity Science, 33, 25184. DOI: 10.17520/biods.2025184.
| 类群 Group | 总计 Total | 经纬度缺失 Missing lat. or log. (%) | 经纬度异常 Abnormal lat. or log. (%) | 高精度数据集 High-resolution dataset (%) | 采样时间异常 Abnormal sampling time (%) | 时间序列数据集 Time series dataset (%) | 全数据集 Full dataset (%) |
|---|---|---|---|---|---|---|---|
| 动物COI Animalia COI | 3,604,970 | 951,853 (26.40%) | 358,576 (9.95%) | 2,294,541 (63.65%) | 169,450 (4.70%) | 2,125,091 (58.95%) | 3,116,067 (86.44%) |
| 植物rbcL Plantae rbcL | 383,112 | 318,694 (83.19%) | 9,041 (2.36%) | 55,377 (14.45%) | 20,279 (5.29%) | 35,098 (9.16%) | 220,722 (57.61%) |
| 真菌ITS Fungi ITS | 2,073,369 | 1,908,945 (92.07%) | 14,574 (0.70%) | 149,850 (7.23%) | 78,940 (3.81%) | 70,910 (3.42%) | 1,318,800 (63.61%) |
表1 动物界、植物界和真菌界的代表性序列统计
Table 1 Statistical analysis of representative sequences across Animalia, Plantae, and Fungi
| 类群 Group | 总计 Total | 经纬度缺失 Missing lat. or log. (%) | 经纬度异常 Abnormal lat. or log. (%) | 高精度数据集 High-resolution dataset (%) | 采样时间异常 Abnormal sampling time (%) | 时间序列数据集 Time series dataset (%) | 全数据集 Full dataset (%) |
|---|---|---|---|---|---|---|---|
| 动物COI Animalia COI | 3,604,970 | 951,853 (26.40%) | 358,576 (9.95%) | 2,294,541 (63.65%) | 169,450 (4.70%) | 2,125,091 (58.95%) | 3,116,067 (86.44%) |
| 植物rbcL Plantae rbcL | 383,112 | 318,694 (83.19%) | 9,041 (2.36%) | 55,377 (14.45%) | 20,279 (5.29%) | 35,098 (9.16%) | 220,722 (57.61%) |
| 真菌ITS Fungi ITS | 2,073,369 | 1,908,945 (92.07%) | 14,574 (0.70%) | 149,850 (7.23%) | 78,940 (3.81%) | 70,910 (3.42%) | 1,318,800 (63.61%) |
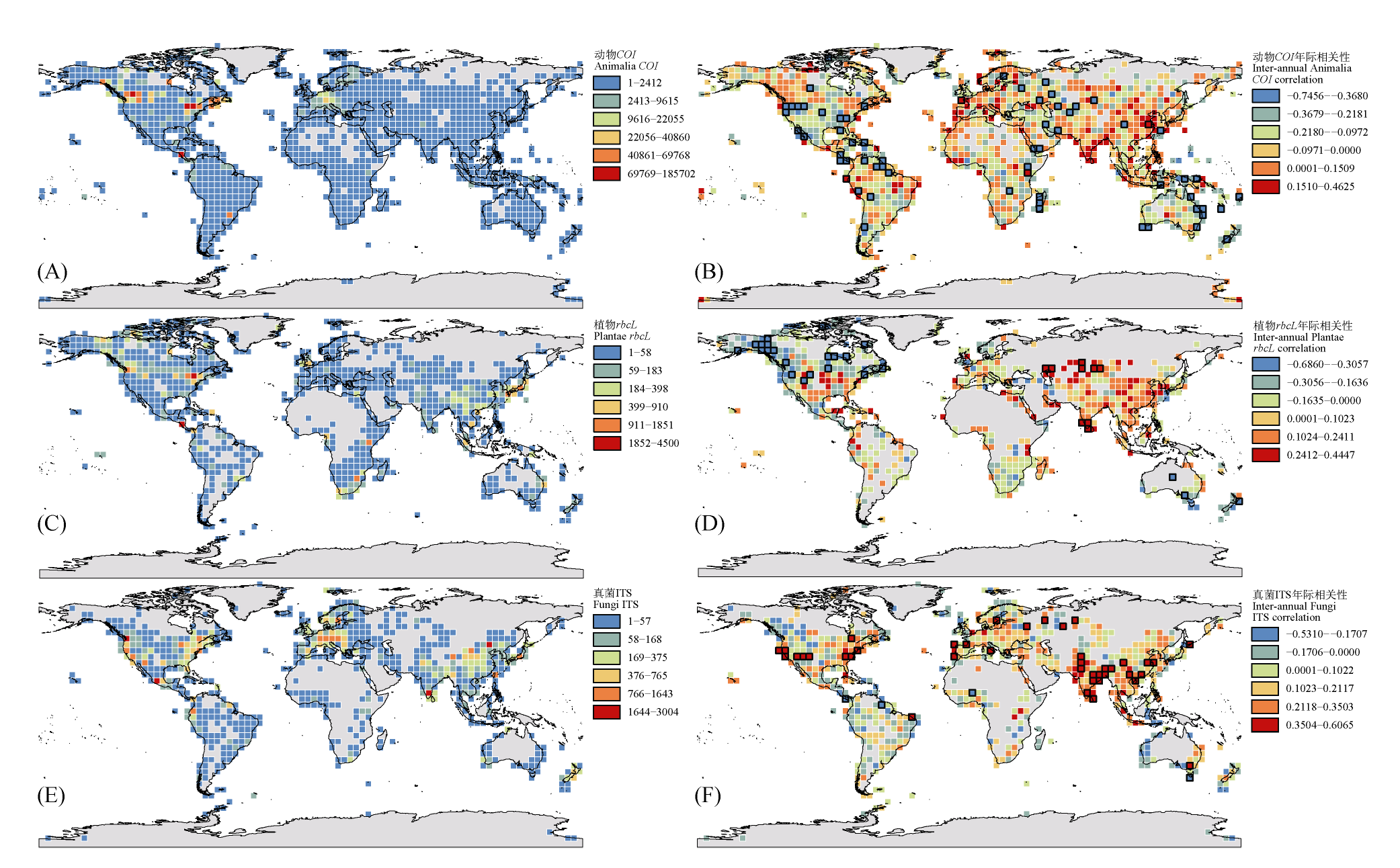
图1 动物界(A、B)、植物界(C、D)和真菌界(E、F)代表序列网格水平全球时空分布格局。左列(A、C、E)分别展示动物界(COI)、植物界(rbcL)和真菌界(ITS)序列的空间分布格局, 基于4° × 4°网格系统, 颜色梯度表示采用自然断点法划分的6个序列密度等级。右列(B、D、F)分别展示动物界(COI)、植物界(rbcL)和真菌界(ITS)序列的年际变化趋势, 通过一般线性模型计算各网格内序列数量与年份的相关系数(正值表示增长趋势, 负值表示下降趋势), 黑色边框表示相关系数达到显著(P < 0.05)的网格。
Fig. 1 Global spatiotemporal distribution patterns of representative sequences at grid level for Animalia (A, B), Plantae (C, D), and Fungi (E, F). The left panels (A, C, E) display the spatial distribution patterns of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS), respectively, based on a 4° × 4° grid system. The color gradient represents six sequence density levels classified using the Jenks natural breaks. The right panels (B, D, F) show the interannual trends of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS), respectively. The correlation coefficient between sequence count and year within each grid was calculated using a general linear model (positive values indicate increasing trends, and negative values indicate decreasing trends). Grids with black borders denote statistically significant correlations (P < 0.05).
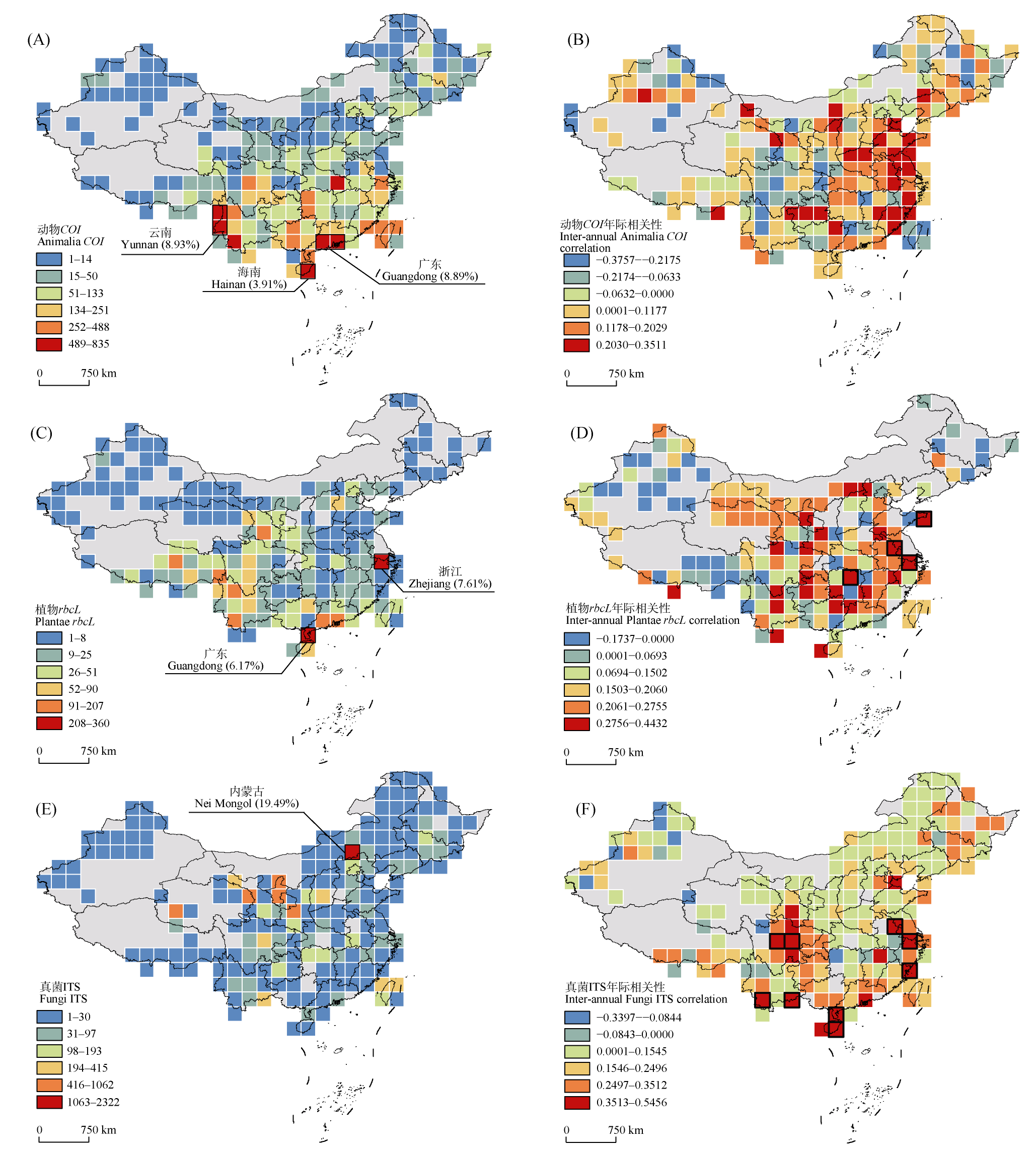
图2 动物界(A、B)、植物界(C、D)和真菌界(E、F)代表序列中国时空分布格局。左列(A、C、E)展示中国区域动物界(COI)、植物界(rbcL)和真菌界(ITS)序列的空间分布格局, 基于2° × 2°网格系统, 颜色梯度表示采用自然断点法划分的6个序列密度等级; 右列(B、D、F)展示中国区域动物界(COI)、植物界(rbcL)和真菌界(ITS)序列的年际变化趋势, 通过一般线性模型计算各网格内序列数量与年份的相关系数(正值表示增长趋势, 负值表示下降趋势), 黑色边框表示相关系数达到显著(P < 0.05)的网格。
Fig. 2 China’s spatiotemporal distribution patterns of representative sequences across Animalia (A, B), Plantae (C, D), and Fungi (E, F) kingdoms. The left panels (A, C, E) display the spatial distribution patterns of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS) within China, based on a 2° × 2° grid system. The color gradient represents six sequence density levels classified using the Jenks natural breaks. The right panels (B, D, F) show the inter-annual trends of sequences from Animalia (COI), Plantae (rbcL), and Fungi (ITS) in China. The correlation coefficient between sequence count and year within each grid was calculated using a general linear model (positive values indicate increasing trends, and negative values indicate decreasing trends). Grids with black borders denote statistically significant correlations (P < 0.05).
| [1] |
Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019) Revisions to the classification, nomenclature, and diversity of eukaryotes. Journal of Eukaryotic Microbiology, 66, 4-119.
DOI PMID |
| [2] | Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Research, 40, D48-D53. |
| [3] |
Boettiger C (2019) Ecological metadata as linked data. Journal of Open Source Software, 4, 1276.
DOI URL |
| [4] |
Boria RA, Olson LE, Goodman SM, Anderson RP (2014) Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecological Modelling, 275, 73-77.
DOI URL |
| [5] |
Brown ED, Williams BK (2019) The potential for citizen science to produce reliable and useful information in ecology. Conservation Biology, 33, 561-569.
DOI PMID |
| [6] |
Chen ZY, Baeza JA, Chen C, Gonzalez MT, González VL, Greve C, Kocot KM, Arbizu PM, Moles J, Schell T, Schwabe E, Sun J, Wong NLWS, Yap-Chiongco M, Sigwart JD (2025) A genome-based phylogeny for Mollusca is concordant with fossils and morphology. Science, 387, 1001-1007.
DOI PMID |
| [7] | Cochrane G, Karsch-Mizrachi I, Takagi T (2016) The International Nucleotide Sequence Database Collaboration. Nucleic Acids Research, 44, D48-D50. |
| [8] | Deck J, Gaither MR, Ewing R, Bird CE, Davies N, Meyer C, Riginos C, Toonen RJ, Crandall ED (2017) The Genomic Observatories Metadatabase (GeOMe): A new repository for field and sampling event metadata associated with genetic samples. PLoS Biology, 15, e2002925. |
| [9] |
Fallon SM (2007) Genetic data and the listing of species under the U.S. Endangered Species Act. Conservation Biology, 21, 1186-1195.
PMID |
| [10] | French CM, Bertola LD, Carnaval AC, Economo EP, Kass JM, Lohman DJ, Marske KA, Meier R, Overcast I, Rominger AJ, Staniczenko PPA, Hickerson MJ (2023) Global determinants of insect mitochondrial genetic diversity. Nature Communications, 14, 5276. |
| [11] | Gaither MR, Bowen BW, Toonen RJ (2013) Population structure in the native range predicts the spread of introduced marine species. Proceedings of the Royal Society B: Biological Sciences, 280, 20130409. |
| [12] |
Gratton P, Marta S, Bocksberger G, Winter M, Trucchi E, Kühl H (2017) A world of sequences: Can we use georeferenced nucleotide databases for a robust automated phylogeography. Journal of Biogeography, 44, 475-486.
DOI URL |
| [13] |
Harris MA, Slippers B, Kemler M, Greve M (2023) Opportunities for diversified usage of metabarcoding data for fungal biogeography through increased metadata quality. Fungal Biology Reviews, 46, 100329.
DOI URL |
| [14] |
Hawksworth DL (2001) The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycological Research, 105, 1422-1432.
DOI URL |
| [15] | Hebert PDN, Cywinska A, Ball SL, Dewaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. |
| [16] |
Hoban S, Bruford M, D’Urban Jackson J, Lopes-Fernandes M, Heuertz M, Hohenlohe PA, Paz-Vinas I, Sjögren-Gulve P, Segelbacher G, Vernesi C, Aitken S, Bertola LD, Bloomer P, Breed M, Rodríguez-Correa H, Funk WC, Grueber CE, Hunter ME, Jaffe R, Liggins L, Mergeay J, Moharrek F, O’Brien D, Ogden R, Palma-Silva C, Pierson J, Ramakrishnan U, Simo-Droissart M, Tani N, Waits L, Laikre L (2020) Genetic diversity targets and indicators in the CBD Post-2020 Global Biodiversity Framework must be improved. Biological Conservation, 248, 108654.
DOI URL |
| [17] | Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim K, Kress WJ, Schneider H, van Alphenstahl J, Barrett SCH, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim Y, Lahaye R, Lee H, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park C, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi D, Little DP (2009) DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA, 106, 12794-12797. |
| [18] | Jenkins GB, Beckerman AP, Bellard C, Benítez López A, Ellison AM, Foote CG, Hufton AL, Lashley MA, Lortie CJ, Ma Z, Moore AJ, Narum SR, Nilsson J, O’Boyle B, Provete DB, Razgour O, Rieseberg L, Riginos C, Santini L, Sibbett B, Peres-Neto PR (2023) Reproducibility in ecology and evolution: Minimum standards for data and code. Ecology and Evolution, 13, 9961. |
| [19] | Jenks GF (1967) The data model concept in statistical mapping. International Yearbook of Cartography, 7, 186-190. |
| [20] |
Mardis ER (2008) Next-generation DNA sequencing methods. Annual Review of Genomics and Human Genetics, 9, 387-402.
DOI PMID |
| [21] |
Meineke EK, Davies TJ, Daru BH, Davis CC (2018) Biological collections for understanding biodiversity in the Anthropocene. Philosophical Transactions of the Royal Society B: Biological Sciences, 374, 20170386.
DOI URL |
| [22] |
Miraldo A, Li S, Borregaard MK, Flórez-Rodríguez A, Gopalakrishnan S, Rizvanovic M, Wang ZH, Rahbek C, Marske KA, Nogués-Bravo D (2016) An Anthropocene map of genetic diversity. Science, 353, 1532-1535.
PMID |
| [23] |
Peng X, Li Q, Cheng ZT, Huang XL (2023) The geography of genetic data: Current status and future perspectives. Frontiers in Ecology and Evolution, 11, 1112636.
DOI URL |
| [24] |
Pope LC, Liggins L, Keyse J, Carvalho SB, Riginos C (2015) Not the time or the place: The missing spatio-temporal link in publicly available genetic data. Molecular Ecology, 24, 3802-3809.
DOI PMID |
| [25] | Powers SM, Hampton SE (2019) Open science, reproducibility, and transparency in ecology. Ecological Applications, 29, e01822. |
| [26] | Ratnasingham S, Hebert PDN (2007) bold: The barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes, 7, 355-364. |
| [27] |
Reddy S (2014) What’s missing from avian global diversification analyses? Molecular Phylogenetics and Evolution, 77, 159-165.
DOI URL |
| [28] | Rhoads A, Au KF (2015) PacBio sequencing and ITS applications. Genomics, Proteomics & Bioinformatics, 13, 278-289. |
| [29] |
Riginos C, Crandall ED, Liggins L, Gaither MR, Ewing RB, Meyer C, Andrews KR, Euclide PT, Titus BM, Therkildsen NO, Salces Castellano A, Stewart LC, Toonen RJ, Deck J (2020) Building a global genomics observatory: Using GEOME (the Genomic Observatories Metadatabase) to expedite and improve deposition and retrieval of genetic data and metadata for biodiversity research. Molecular Ecology Resources, 20, 1458-1469.
DOI PMID |
| [30] | Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Consortium FB (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences, USA, 109, 6241-6246. |
| [31] |
Tannenbaum C, Ellis RP, Eyssel F, Zou J, Schiebinger L (2019) Sex and gender analysis improves science and engineering. Nature, 575, 137-146.
DOI |
| [32] |
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology, 31, 21-32.
DOI PMID |
| [33] |
Troudet J, Grandcolas P, Blin A, Vignes-Lebbe R, Legendre F (2017) Taxonomic bias in biodiversity data and societal preferences. Scientific Reports, 7, 9132.
DOI PMID |
| [34] |
Vines TH, Andrew RL, Bock DG, Franklin MT, Gilbert KJ, Kane NC, Moore JS, Moyers BT, Renaut S, Rennison DJ, Veen T, Yeaman S (2013) Mandated data archiving greatly improves access to research data. The Federation of American Societies for Experimental Biology Journal, 27, 1304-1308.
DOI URL |
| [35] |
Warnasuriya SD, Udayanga D, Manamgoda DS, Biles C (2023) Fungi as environmental bioindicators. Science of the Total Environment, 892, 164583.
DOI URL |
| [36] |
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J, Da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B (2016) The FAIR guiding principles for scientific data management and stewardship. Scientific Data, 3, 160018.
DOI |
| [37] |
Winter DJ (2017) rentrez: An R package for the NCBI eUtils API. The R Journal, 9, 520.
DOI URL |
| [38] | Wood DA, Vandergast AG, Barr KR, Inman RD, Esque TC, Nussear KE, Fisher RNBM (2013) Comparative phylogeography reveals deep lineages and regional evolutionary hotspots in the Mojave and Sonoran Deserts. Diversity & Distributions, 19, 722-737. |
| [39] |
Yan DF, Mills JG, Gellie NJC, Bissett A, Lowe AJ, Breed MF (2018) High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biological Conservation, 217, 113-120.
DOI URL |
| [40] |
Yao G, Zhang YQ, Barrett C, Xue B, Bellot S, Baker WJ, Ge XJ (2023) A plastid phylogenomic framework for the palm family (Arecaceae). BMC Biology, 21, 50.
DOI PMID |
| [41] |
Zizka A, Silvestro D, Andermann T, Azevedo J, Ritter CD, Edler D, Farooq H, Herdean A, Ariza M, Scharn R, Svantesson S, Wengström N, Zizka V, Antonelli A, Quental T (2019) CoordinateCleaner: Standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution, 10, 744-751.
DOI URL |
| [1] | 洪欣艺, 蔡易朗, 方嘉乐, 姚可侃, 李佳乐, 王懿祥, 白尚斌, 王楠, 周秀梅. 西溪湿地土壤病毒多样性及其碳代谢基因解析[J]. 生物多样性, 2025, 33(9): 25190-. |
| [2] | 旦增尼玛, 孙伟, 李聪, 张纾意, 赵竹楠, 许永强, 普布卓玛, 罗诗琦, 达娃, 周欣. 西藏吉隆沟地区开花植物叶绿体基因组数据集[J]. 生物多样性, 2025, 33(9): 25270-. |
| [3] | 薛瑞翔, 马雪蓉, 吴炯文, 刘爱君, 张细权, 季从亮, 殷颖珊, 朱炜健, 罗庆斌. 中山麻鸭群体遗传多样性与遗传结构[J]. 生物多样性, 2025, 33(8): 24592-. |
| [4] | 王凤英, 吴增源, 崔涵, 李垠蕾, 邓莉娟, 王红, 刘杰. 第三极荨麻属麻叶荨麻分支的物种界限[J]. 生物多样性, 2025, 33(8): 25138-. |
| [5] | 李慧霞, 李玉, 宁馨, 李晓晨, 王天瑞, 宋以刚, 戴锡玲, 郑斯斯, 钟鑫. 基于叶绿体基因组的江南牡丹草遗传多样性与遗传结构[J]. 生物多样性, 2025, 33(8): 25149-. |
| [6] | 虎灵, 沈泽昊. 地理基因组学: 研究方法与进展[J]. 生物多样性, 2025, 33(7): 25010-. |
| [7] | 卢晓强, 董姗姗, 马月, 徐徐, 邱凤, 臧明月, 万雅琼, 李孪鑫, 于赐刚, 刘燕. 前沿技术在生物多样性研究中的应用现状、挑战与展望[J]. 生物多样性, 2025, 33(4): 24440-. |
| [8] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [9] | 曹东, 李焕龙, 彭扬, 魏存争. 植物基因组大小与性状关系的研究进展[J]. 生物多样性, 2025, 33(2): 24192-. |
| [10] | 张奕涵, 杨光, 周青松, 牛泽清, 朱朝东, 罗阿蓉. 蜜蜂类昆虫系统发生基因组学研究概况[J]. 生物多样性, 2025, 33(10): 25234-. |
| [11] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [12] | 姚祥坦, 张心怡, 陈阳, 袁晔, 程旺大, 王天瑞, 邱英雄. 基于基因组重测序揭示栽培欧菱遗传多样性及‘南湖菱’的起源驯化历史[J]. 生物多样性, 2024, 32(9): 24212-. |
| [13] | 罗小燕, 李强, 黄晓磊. 戴云山国家级自然保护区访花昆虫DNA条形码数据集[J]. 生物多样性, 2023, 31(8): 23236-. |
| [14] | 邢超, 林依, 周智强, 赵联军, 蒋仕伟, 林蓁蓁, 徐基良, 詹祥江. 基于DNA条形码技术构建王朗国家级自然保护区陆生脊椎动物遗传资源数据库及物种鉴定[J]. 生物多样性, 2023, 31(7): 22661-. |
| [15] | 吴帆, 刘深云, 江虎强, 王茜, 陈开威, 李红亮. 中华蜜蜂和意大利蜜蜂秋冬期传粉植物多样性比较[J]. 生物多样性, 2023, 31(5): 22528-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn