



生物多样性 ›› 2010, Vol. 18 ›› Issue (5): 461-472. DOI: 10.3724/SP.J.1003.2010.461 cstr: 32101.14.SP.J.1003.2010.461
所属专题: 生物多样性信息学专题(I)
收稿日期:2010-01-19
接受日期:2010-06-11
出版日期:2010-09-20
发布日期:2010-09-20
通讯作者:
纪力强
作者简介:* E-mail: ji@ioz.ac.cn基金资助:
Congtian Lin1,2, Liqiang Ji1,*( )
)
Received:2010-01-19
Accepted:2010-06-11
Online:2010-09-20
Published:2010-09-20
Contact:
Liqiang Ji
摘要:
根据对生物分布地预测模型和软件发展现状的分析和总结, 本研究在PSDS 1.0的基础上提出并实现一个基于GIS且具有多个代表性模型的生物分布地预测系统(PSDS 2.0)。PSDS 2.0系统继承了1.0的环境包络和聚类包络模型, 进一步引入了限制因子包络、马氏距离、支持向量机等新模型, 并针对本领域中模型比较与选择的难点增加了迭代交叉验证的多模型选择功能。系统还实现了灵活定制和评估伪负样本的功能, 通过用只需要正样本的I类模型预测的结果对随机产生的伪负样本进行评估, 减小其落入适宜地区的概率, 进一步提高需要正负样本的II类模型的准确率。GIS功能在PSDS 2.0中也得到加强, 被应用于数据准备及结果分析等重要环节。文章最后以白冠长尾雉(Syrmaticus reevesii)为例, 运用PSDS 2.0系统预测其在中国范围内的潜在分布地, 并对各种模型的预测结果进行评估和比较。
林聪田, 纪力强 (2010) PSDS 2.0: 一个基于GIS和多个模型的生物潜在分布地预测系统. 生物多样性, 18, 461-472. DOI: 10.3724/SP.J.1003.2010.461.
Congtian Lin, Liqiang Ji (2010) PSDS (predictive species distribution system) 2.0: a system based on GIS and multiple models for predicting potential distribution of species. Biodiversity Science, 18, 461-472. DOI: 10.3724/SP.J.1003.2010.461.
| 软件 Software | GIS功能 With GIS | 模型算法 Algorithm | 分布数据类型 Distribution type | 变量类型 Variable type | 预测结果 Result type | 软件来源和重要文献 Software source/Main references |
|---|---|---|---|---|---|---|
| Desktop GARP | + | 预设规则遗传算法 Genetic Algorithm for Rule-set Production(GARP) | P | Co/Ca | B | desktopgarp/Download.html |
| DOMAIN | + | Gower Metric (GM) | P | Co/Ca | Si | Knowledge/Toolkits/domain |
| DIVA-GIS | + | 环境包络 Environment Envelope (EE) | P | Co | B | |
| Gower Metric距离 Gower Metric Distance (GM) | P | Co/Ca | Si | |||
| CLIMEX | + | 生态气候指数 Ecological Index (EI) | P | Co | B | No free link |
| PSDS 1.0 | + | 环境包络 Environment Envelope (EE) | P | Co | B | ji@ioz.ac.cn或linct@ioz.ac.cn |
| 聚类包络 Hierarchical Cluster plus Environmental Envelope (HCEM) | P | Co | B | |||
| Biomapper | + | 生态位因子分析 Ecological Niche Factor Analysis (ENFA) | P | Co | Su | products.html |
| MaxEnt | - | 最大熵模型 Maximum Entropy (MaxEnt) | P | Co | Pro | ~schapire/maxent |
| GeoSVM | - | 支持向量机 Support Vector Machine (SVM) | P & A | Co/Ca | B | |
| Neuroet | - | 人工神经网络 Artificial Neural Network (ANN) | P & A | Co/Ca | B | |
| SPECIES | - | 人工神经网络 Artificial Neural Network (ANN) | P & A | Co/Ca | B | |
| Agroclimatic Library | - | 农业气候相似距 Agricultural Similar Distance (ASD) | P | Co | Si | No open link |
| Random Forest | - | 随机森林 Random Forest (RF) | P & A | Co/Ca | B |
表1 常用的生物分布地预测软件与模型
Table 1 General description of the software applications and models for predicting species’ distribution
| 软件 Software | GIS功能 With GIS | 模型算法 Algorithm | 分布数据类型 Distribution type | 变量类型 Variable type | 预测结果 Result type | 软件来源和重要文献 Software source/Main references |
|---|---|---|---|---|---|---|
| Desktop GARP | + | 预设规则遗传算法 Genetic Algorithm for Rule-set Production(GARP) | P | Co/Ca | B | desktopgarp/Download.html |
| DOMAIN | + | Gower Metric (GM) | P | Co/Ca | Si | Knowledge/Toolkits/domain |
| DIVA-GIS | + | 环境包络 Environment Envelope (EE) | P | Co | B | |
| Gower Metric距离 Gower Metric Distance (GM) | P | Co/Ca | Si | |||
| CLIMEX | + | 生态气候指数 Ecological Index (EI) | P | Co | B | No free link |
| PSDS 1.0 | + | 环境包络 Environment Envelope (EE) | P | Co | B | ji@ioz.ac.cn或linct@ioz.ac.cn |
| 聚类包络 Hierarchical Cluster plus Environmental Envelope (HCEM) | P | Co | B | |||
| Biomapper | + | 生态位因子分析 Ecological Niche Factor Analysis (ENFA) | P | Co | Su | products.html |
| MaxEnt | - | 最大熵模型 Maximum Entropy (MaxEnt) | P | Co | Pro | ~schapire/maxent |
| GeoSVM | - | 支持向量机 Support Vector Machine (SVM) | P & A | Co/Ca | B | |
| Neuroet | - | 人工神经网络 Artificial Neural Network (ANN) | P & A | Co/Ca | B | |
| SPECIES | - | 人工神经网络 Artificial Neural Network (ANN) | P & A | Co/Ca | B | |
| Agroclimatic Library | - | 农业气候相似距 Agricultural Similar Distance (ASD) | P | Co | Si | No open link |
| Random Forest | - | 随机森林 Random Forest (RF) | P & A | Co/Ca | B |
| 生物气候因子 Bioclimatic factors | 因子分析 Factor analysis | KMO | Bartlett’s 检验 Bartlett’s test | 新的因子 New factors |
|---|---|---|---|---|
| 平均月温差 Mean diurnal range 年平均湿度 Annual mean humidity 年降水 Annual precipitation 年平均温度 Annual mean temperature 降水的季节变化因子 Precipitation seasonality 温度的季节变化因子 Temperature seasonality 等温因子 Isothermality 年温差 Temperature annual range | 因子提取方法: 主成分分析; 旋转方法: Varimax with Kaiser Normalization. Extracted method: Principal Component Analysis. Rotation Method: Varimax with Kaiser Normalization. | 0.665 | Approx. Chi-Square =7,046,789.32 df=28 Sig.=0.000 | 平均生物气候因子 Mean bioclimatic factor |
| 季节性生物气候因子 Seasonal bioclimatic factor | ||||
| 霜降月平均降水 Mean precipitation of frost free month 最湿润季节的降水 Precipitation of wettest quarter 最湿润月份的降水 Precipitation of wettest month 最温暖月份的降水 Precipitation of warmest quarter 最干燥月份的降水 Precipitation of driest quarter 最冷季节的降水 Precipitation of coldest quarter 最干燥月份的降水 Precipitation of driest month 最冷月份的最低温度 Min temperature of coldest month 最冷季节的平均温度 Mean temperature of coldest quarter 最暖月的最高温度 Max temperature of warmest month 最暖季的平均温度 Mean temperature of warmest quarter 最湿润季节的平均温度 Mean temperature of wettest quarter 霜降月的平均温度 Mean temperature of frost free month 平均霜降天数 Mean frost days 最干燥季节的平均温度 Mean temperature of driest quarter | 同上 Same as above | 0.875 | Approx. Chi-Square =25,734,237.85 df=105 Sig.=0.000 | 与降水相关的极端生物气候因子Harsh bioclimatic factor related to precipitation |
| 与温度相关的极端生物气候因子Harsh bioclimatic factor related to temperature |
表2 对23个生物气候变量进行因子分析和显著性检验
Table 2 The result of factor analysis of the bioclimatic variables and significance test
| 生物气候因子 Bioclimatic factors | 因子分析 Factor analysis | KMO | Bartlett’s 检验 Bartlett’s test | 新的因子 New factors |
|---|---|---|---|---|
| 平均月温差 Mean diurnal range 年平均湿度 Annual mean humidity 年降水 Annual precipitation 年平均温度 Annual mean temperature 降水的季节变化因子 Precipitation seasonality 温度的季节变化因子 Temperature seasonality 等温因子 Isothermality 年温差 Temperature annual range | 因子提取方法: 主成分分析; 旋转方法: Varimax with Kaiser Normalization. Extracted method: Principal Component Analysis. Rotation Method: Varimax with Kaiser Normalization. | 0.665 | Approx. Chi-Square =7,046,789.32 df=28 Sig.=0.000 | 平均生物气候因子 Mean bioclimatic factor |
| 季节性生物气候因子 Seasonal bioclimatic factor | ||||
| 霜降月平均降水 Mean precipitation of frost free month 最湿润季节的降水 Precipitation of wettest quarter 最湿润月份的降水 Precipitation of wettest month 最温暖月份的降水 Precipitation of warmest quarter 最干燥月份的降水 Precipitation of driest quarter 最冷季节的降水 Precipitation of coldest quarter 最干燥月份的降水 Precipitation of driest month 最冷月份的最低温度 Min temperature of coldest month 最冷季节的平均温度 Mean temperature of coldest quarter 最暖月的最高温度 Max temperature of warmest month 最暖季的平均温度 Mean temperature of warmest quarter 最湿润季节的平均温度 Mean temperature of wettest quarter 霜降月的平均温度 Mean temperature of frost free month 平均霜降天数 Mean frost days 最干燥季节的平均温度 Mean temperature of driest quarter | 同上 Same as above | 0.875 | Approx. Chi-Square =25,734,237.85 df=105 Sig.=0.000 | 与降水相关的极端生物气候因子Harsh bioclimatic factor related to precipitation |
| 与温度相关的极端生物气候因子Harsh bioclimatic factor related to temperature |
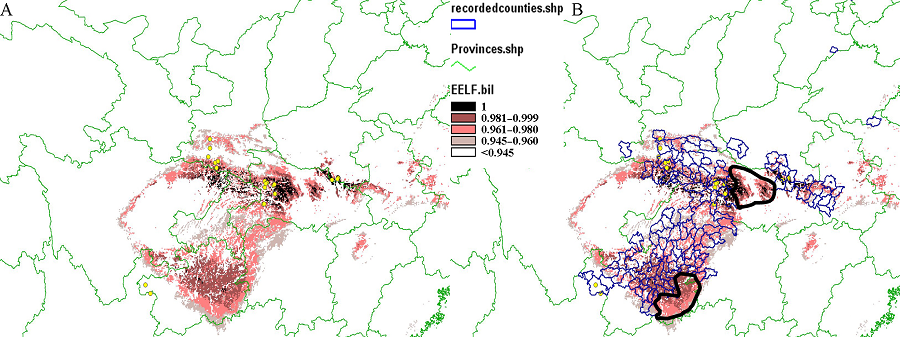
图3 限制因子模型预测结果图(A)及与分布记录的县级图进行叠加分析的结果(B)。可以看出大部分预测到的区域与历史记录的地区相吻合。图中两个黑色多边形是预测到适宜白冠长尾雉分布但无文献记录的区域, 上方的黑色多边形区域处于湖北省中西部地区, 具有比较高的适宜度, 靠下方的黑色多边形区域处于贵州省东部, 是白冠长尾雉的近缘种白颈长尾雉(Syrmaticus ellioti)的分布地。
Fig. 3 The result map shows the suitable area predicted by the EELF model in the relative part of China (A) and the overlay analysis of the EELF result with the map of recorded counties (B). The most area predicted to be suitable is similar to the recorded area. The sites outlined by the two black polygons are predicted to be suitable for target species without historical records. The area in the upper polygon is in the middle to west of Hubei Province with very high suitability but without records. The area in the other polygon in the east of Guizhou Province is the distribution area of the relative species Syrmaticus ellioti.
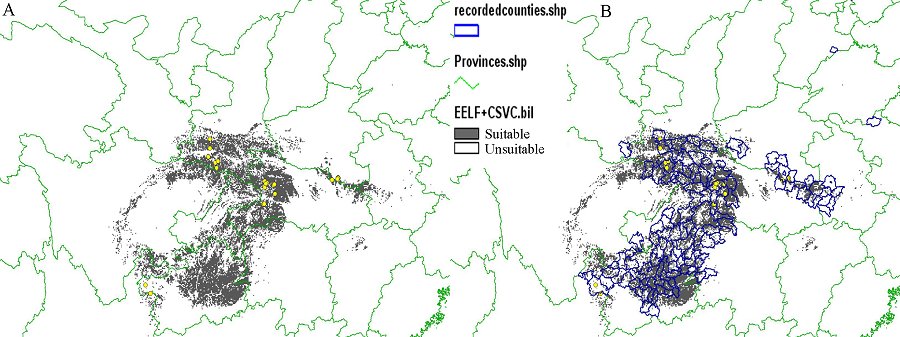
图4 EELF + C-SVC模型预测结果图(A)及与分布记录的县级图进行叠加分析的结果(B), 大部分预测到的区域与历史记录的区域相吻合。
Fig. 4 The result map from EELF pseudo-absences + C-SVC (A) and the overlay analysis of the result map with the map of recorded counties (B). The most area predicted to be suitable for target species is similar to the recorded area.
| [1] | Arif S, Adams DC, Jill AW (2007) Bioclimatic modelling, morphology, and behaviour reveal alternative mechanisms regulating the distributions of two parapatric salamander species. Evolutionary Ecology Research, 9, 843-854. |
| [2] | BirdLife International (2001) Threatened Birds of Asia: the BirdLife International Red Data. BirdLife International, Cambridge, UK. |
| [3] | Breiman (2001) Random forest. Machine Learning, 45, 5-32. |
| [4] | Busby JR (1991) BIOCLIM: a bioclimatic analysis and prediction system. In: Nature Conservation: Cost Effective Biological Surveys and Data Analysis (eds Margules CR, Austin MP), pp. 64-68. Commonwealth Scientific and Industrial Research Organization, Canberra, Australia. |
| [5] | Carpenter G, Gillison AN, Winter J (1993) DOMAIN: a flexible modeling procedure for mapping potential distributions of plants and animals. Biodiversity and Conservation, 2, 667-680. |
| [6] | Chang Chih-Chung, Lin Chih-Jen (2001) LIBSVM: a library for support vector machines. http://www.csie.ntu.edu.tw/~cjlin/libsvm |
| [7] | Chen GJ, Peterson AT (2002) Prioritization of areas in China for the conservation of endangered birds using modeled geographical distributions. Bird Conservation International, 12, 197-209. |
| [8] | Deng H (邓浩), Ji LQ (纪力强) (2008) Design and implementation of PSDS system, a software for predicting the potential distribution of species. Biodiversity Science (生物多样性), 16, 96-102. (in Chinese with English abstract) |
| [9] | Ding P (丁平) (1998) The distributions and systematic analysis of the long-tailed pheasants. Life Science Research (生命科学研究), 2, 122-131. (in Chinese with English abstract) |
| [10] | Drake JM, Randin C, Guisan A (2006) Modelling ecological niches with support vector machines. Journal of Applied Ecology, 43, 424-432. |
| [11] | Fawcett T (2006) An introduction to ROC analysis. Pattern Recognition Letters, 27, 861-874. |
| [12] | Guisan A, Zimmermann NE (2000) Predictive habitat distribution models in ecology. Ecological Modelling, 135, 147-186. |
| [13] | Guisan A, Stuart BW, Andrew DW (1999) GLM versus CCA spatial modeling of plant species distribution. Plant Ecology, 143, 107-122. |
| [14] | Guo QH, Kelly M, Graham CH (2005) Support vector machines for predicting distribution of Sudden Oak Death in California. Ecological Modelling, 182, 75-90. |
| [15] | Hirzel AH, Hausser J, Perrin N (2002) Ecological-niche factor analysis: how to compute habitat suitability maps without absence data? Ecology, 83, 2027-2036. |
| [16] | Hou BH (侯柏华), Zhang RJ (张润杰) (2005) Potential distributions of the fruit fly Bactrocera dorsalis (Diptera: Tephritidae) in China as predicted by CLIMEX. Acta Ecologica Sinica (生态学报), 25, 1569-1574. (in Chinese with English abstract) |
| [17] | Irfan-Ullah M, Amarnath G, Murthy MSR, Peterson AT (2007) Mapping the geographic distribution of Aglaia bourdillonii Gamble (Meliaceae), an endemic and threatened plant, using ecological niche modeling. Biodiversity and Conservation, 16, 1917-1925. |
| [18] | John M (马敬能), Karen P (菲利普斯), He FQ (何芬奇) (2000) A Field Guide to the Birds of China (中国鸟类野外手册). Hunan Education Press, Changsha. (in Chinese) |
| [19] | Levin R (1966) The strategy of model building in population ecology. American Scientist, 54, 421-431. |
| [20] | Li J, Hilbert DW (2007) LIVES: a new habitat modeling technique for predicting the distribution of species’ occurrences using presence-only data based on limiting factor theory. Biodiversity and Conservation, 17, 1-17. |
| [21] | Li HM (李红梅), Han HX (韩红香), Xue DY (薛大勇) (2005) Prediction of potential geographic distribution areas for the pine bark scale, Matsucoccus matsumurae (Kuwana) (Homoptera: Margarodidae) in China using GARP modeling system. Acta Entomologica Sinica (昆虫学报), 48, 95-100. (in Chinese with English abstract) |
| [22] | Lobo MJ, Jimenez-Valverde A, Real R (2008) AUC: a misleading measure of the performance of predictive distribution models. Global Ecology and Biogeography, 17, 145-151. |
| [23] | Lu TC (卢汰春), Zhang WF (张万福) (1993) Rare and Endangered Birds of China: Life History and Nurse of Phasianidae and Tetraonidae (中国珍稀濒危鸟类: 雉科、松鸡科鸟类生活史与保育). China Taiwan Science and Technology Press, Taizhong. (in Chinese) |
| [24] | Muñoz MES, Giovanni R, Siqueira MF, Sutton T, Brewer P, Pereira RS, Canhos DAL, Canhos VP (2009) openModeller: a generic approach to species’ potential distribution modelling. GeoInformatica, DOI: 10.1007/s10707-009-0090-7. |
| [25] | Mahalanobis PC (1930) On tests and measures of group divergence. Asiatic Society of Bengal, 26, 541-588. |
| [26] | New M, Lister D, Hulme M, Makin I (2002) A high-resolution data set of surface climate over global land areas. Climate Research, 21, 1-25. |
| [27] | Noble PA, Tribou EH (2007) Neuroet: an easy-to-use artificial neural network for ecological and biological modeling. Ecological Modelling, 203, 87-98. |
| [28] | Pearson RG, Dawson TP (2003) Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecology & Biogeography, 12, 361-371. |
| [29] | Pearson RG, Dawson TP, Berry PM, Harrison PA (2002) SPECIES: a spatial evaluation of climate impact on the envelope of species. Ecological Modelling, 154, 289-300. |
| [30] | Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190, 231-259. |
| [31] | Robert JH, Susan EC, Juan LP, Peter GJ (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978. |
| [32] | Rotenberry JT, Preston KL, Knick ST (2006) GIS-based niche modeling for mapping species’ habitat. Ecology, 6, 1458-1464. |
| [33] | Schröder B (2004) ROC-AUC: ROC Plotting and AUC Calculation Transferability test. http://brandenburg.geoecology.uni-potsdam.de/users/schroeder/download.html. |
| [34] | Seymour G (1993) Predictive Inference. Chapman and Hall, New York. |
| [35] | Stockwell D, Peters D (1999) The GARP modeling system: problems and solutions to automated spatial prediction. International Journal of Geographic Information System, 13, 143-158. |
| [36] |
Scholköpf B, Platt J, Shawe-Taylor J, Smola AJ, Williamson RC (2001) Estimating the support of a high-dimensional distribution. Neural Computation, 13, 1443-1471.
DOI URL PMID |
| [37] | Sutton T, Giovanni R, Siqueira MF (2007) Introducing openModeller. OSGEEO Journal, 1, 1-5. |
| [38] | The U.S. Geological Survey (2005) Global 30-Arc-Second Elevation Data Set. http://www.usgs.gov. |
| [39] | Vapnik (1995) The Nature of Statistical Learning Theory. Springer-Verlag, New York. |
| [40] | Wang XJ (汪兴鉴), Huang DC (黄顶成), Li HM (李红梅), Xue DY (薛大勇), Zhang RZ (张润志), Chen XL (陈小琳) (2006) Invasion and identification of Liriomyza trifolii and its potential distribution areas in China. Chinese Bulletin of Entomology (昆虫知识), 43, 540-545. (in Chinese with English abstract) |
| [41] | Wildlife Conservation (WCS) and Center for International Earth Science Information Network (CIESIN) (2005) Last of the Wild Data Version 2, 2005(LTW-2): Global Human Footprint Dataset (Geographic). http://sedac.ciesin.columbia.edu/wildareas/ |
| [42] | Wu ZK (吴至康) (1986) Guizhou Avifauna (贵州鸟类志). Guizhou People’s Publishing House, Guiyang. (in Chinese) |
| [43] | Yang L (杨岚) (1994) Yunnan Avifauna (云南鸟类志). Yunnan Science and Technology Publishing House, Kunming. (in Chinese) |
| [44] | Zuo WY, Lao N, Geng YY, Ma KP (2008) GeoSVM: an efficient and effective tool to predict species’ potential distributions. Journal of Plant Ecology, 1, 143-145. |
| [45] | Zhuo WH (卓卫华), Yin SJ (阴三军), Li LJ (李灵军) (2000) Illustrated Handbook of Protected Wild Animals in Henan (河南保护野生动物图鉴). Yellow River Irrigation Press, Zhengzhou. (in Chinese) |
| [46] | Zheng SW (郑生武) (1994) Fauna of Rare and Endangered Animals in Northwestern China (中国西北地区珍稀动物志). China Forestry Publishing House, Beijing. (in Chinese) |
| [1] | 刘志发, 王新财, 龚粤宁, 陈道剑, 张强. 基于红外相机监测的广东南岭国家级自然保护区鸟兽多样性及其垂直分布特征[J]. 生物多样性, 2023, 31(8): 22689-. |
| [2] | 刘向, 张鹏, 刘建全. 无机肥料是青海塔拉滩光伏电站植被恢复过程中的限制性因子[J]. 生物多样性, 2022, 30(5): 22100-. |
| [3] | 王然, 乔慧捷. 生态位模型在流行病学中的应用[J]. 生物多样性, 2020, 28(5): 579-586. |
| [4] | 范靖宇, 李汉芃, 杨琢, 朱耿平. 基于本土最优模型模拟入侵物种水盾草在中国的潜在分布[J]. 生物多样性, 2019, 27(2): 140-148. |
| [5] | 丁晨晨, 胡一鸣, 李春旺, 蒋志刚. 印度野牛在中国的分布及其栖息地适宜性分析[J]. 生物多样性, 2018, 26(9): 951-961. |
| [6] | 王波, 黄勇, 李家堂, 戴强, 王跃招, 杨道德. 西南喀斯特地貌区两栖动物丰富度分布格局与环境因子的关系[J]. 生物多样性, 2018, 26(9): 941-950. |
| [7] | 周中一, 刘冉, 时书纳, 苏艳军, 李文楷, 郭庆华. 基于激光雷达数据的物种分布模拟: 以美国加州内华达山脉南部区域食鱼貂分布模拟为例[J]. 生物多样性, 2018, 26(8): 878-891. |
| [8] | 叶俊伟, 袁永革, 蔡荔, 王晓娟. 中国东北温带针阔混交林植物物种的谱系地理研究进展[J]. 生物多样性, 2017, 25(12): 1339-1349. |
| [9] | 方晓峰, 杨庆松, 刘何铭, 马遵平, 董舒, 曹烨, 袁铭皎, 费希旸, 孙小颖, 王希华. 天童常绿阔叶林中常绿与落叶物种的物种多度分布格局[J]. 生物多样性, 2016, 24(6): 629-638. |
| [10] | 朱耿平, 乔慧捷. Maxent模型复杂度对物种潜在分布区预测的影响[J]. 生物多样性, 2016, 24(10): 1189-1196. |
| [11] | 崔相艳, 王文娟, 杨小强, 李述, 秦声远, 戎俊. 基于生态位模型预测野生油茶的潜在分布[J]. 生物多样性, 2016, 24(10): 1117-1128. |
| [12] | 乔慧捷, 林聪田, 王江宁, 纪力强. 流程化的生态建模方法与科学工作流系统[J]. 生物多样性, 2014, 22(3): 277-284. |
| [13] | 朱耿平, 刘强, 高玉葆. 提高生态位模型转移能力来模拟入侵物种 的潜在分布[J]. 生物多样性, 2014, 22(2): 223-230. |
| [14] | 朱耿平, 刘国卿, 卜文俊, 高玉葆. 生态位模型的基本原理及其在生物多样性保护中的应用[J]. 生物多样性, 2013, 21(1): 90-98. |
| [15] | 陈立立, 余岩, 何兴金. 喜旱莲子草在中国的入侵和扩散动态及其潜在分布区预测[J]. 生物多样性, 2008, 16(6): 578-585. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn