



生物多样性 ›› 2009, Vol. 17 ›› Issue (5): 476-481. DOI: 10.3724/SP.J.1003.2009.09100 cstr: 32101.14.SP.J.1003.2009.09100
收稿日期:2009-04-16
接受日期:2009-09-05
出版日期:2009-09-20
发布日期:2009-09-20
通讯作者:
朱华
作者简介:*E-mail: zhuh@xtbg.ac.cn基金资助:
Longqian Xiao1,2, Hua Zhu1,*( )
)
Received:2009-04-16
Accepted:2009-09-05
Online:2009-09-20
Published:2009-09-20
Contact:
Hua Zhu
摘要:
本研究对苏铁(Cycas revoluta) nrDNA ITS进行克隆测序, 并以cDNA ITS为参照, 比较分析获得的序列的碱基变异、GC含量、5.8S二级结构的稳定性和5.8S保守基序的有无以及系统发育关系。结果发现苏铁nrDNA ITS存在较高的基因组内多样性, 同时, 这些分化的nrDNA ITS拷贝中包含有假基因的存在, 而且假基因与功能拷贝之间已经形成了较大的遗传分化, 这暗示假基因起源有较长历史。苏铁核仁组织区不仅多达16个, 而且分布在13条染色体上, 这可能是其nrDNA ITS致同进化不完全的主要原因。
肖龙骞, 朱华 (2009) 苏铁nrDNA ITS区的序列多态性:不完全致同进化的证据. 生物多样性, 17, 476-481. DOI: 10.3724/SP.J.1003.2009.09100.
Longqian Xiao, Hua Zhu (2009) Intra-genomic polymorphism in the internal transcribed spacer (ITS) regions of Cycas revoluta: evidence of incomplete concerted evolution. Biodiversity Science, 17, 476-481. DOI: 10.3724/SP.J.1003.2009.09100.
| 克隆编号 Clone No. | GenBank序列号 GenBank accession no. | 长度(GC含量%) Length (bp) (GC content %) | 5.8S基序5.8S motif | 最小自由能5.8S Δ G (kcal/mol) | ||
|---|---|---|---|---|---|---|
| ITS1 | ITS2 | 5.8S | ||||
| D1 | FJ907980 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D2 | FJ907972 | 677(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D3 | FJ908060 | 674(51.0) | 243(46.1) | 161(44.7) | V | -13.79 |
| D4 | FJ907973 | 677(63.8) | 246(65.9) | 161(55.9) | V | -16.39 |
| D5 | FJ907974 | 677(64.1) | 246(65.0) | 161(55.9) | V | -16.39 |
| D6 | FJ908059 | 676(49.4) | 243(49.4) | 161(43.5) | V | -8.54 |
| D7 | FJ908058 | 598(49.3) | 243(46.1) | 160(43.1) | -11.51 | |
| D8 | FJ908051 | 672(51.0) | 243(50.6) | 161(37.9) | -10.95 | |
| D9 | FJ907982 | 676(63.8) | 246(65.0) | 161(55.9) | V | -16.39 |
| D11 | FJ908056 | 679(50.5) | 242(50.0) | 161(42.9) | -9.42 | |
| D12 | FJ907981 | 677(64.0) | 246(65.0) | 161(55.3) | V | -16.90 |
| D13 | FJ907983 | 674(62.7) | 246(62.6) | 161(54.7) | V | -19.39 |
| D14 | FJ908045 | 658(50.2) | 243(50.6) | 161(43.5) | -10.37 | |
| D15 | FJ908067 | 671(48.7) | 243(50.2) | 161(44.7) | V | -13.21 |
| C1 | FJ907976 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| C2 | FJ907979 | 678 (64.0) | 246(65.0) | 161(55.9) | V | -16.39 |
| C3 | FJ907978 | 679(64.1) | 246(65.0) | 161(55.3) | V | -16.59 |
| C4 | FJ907977 | 677 (64.1) | 246(64.6) | 161(55.9) | V | -16.39 |
| C5 | FJ907975 | 681(64.2) | 230(65.6) | 161(55.9) | V | -16.39 |
表1 苏铁中ITS1、5.8S和ITS2的特征
Table 1 The characteristics of ITS1, 5.8S and ITS2 in Cycas revoluta
| 克隆编号 Clone No. | GenBank序列号 GenBank accession no. | 长度(GC含量%) Length (bp) (GC content %) | 5.8S基序5.8S motif | 最小自由能5.8S Δ G (kcal/mol) | ||
|---|---|---|---|---|---|---|
| ITS1 | ITS2 | 5.8S | ||||
| D1 | FJ907980 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D2 | FJ907972 | 677(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| D3 | FJ908060 | 674(51.0) | 243(46.1) | 161(44.7) | V | -13.79 |
| D4 | FJ907973 | 677(63.8) | 246(65.9) | 161(55.9) | V | -16.39 |
| D5 | FJ907974 | 677(64.1) | 246(65.0) | 161(55.9) | V | -16.39 |
| D6 | FJ908059 | 676(49.4) | 243(49.4) | 161(43.5) | V | -8.54 |
| D7 | FJ908058 | 598(49.3) | 243(46.1) | 160(43.1) | -11.51 | |
| D8 | FJ908051 | 672(51.0) | 243(50.6) | 161(37.9) | -10.95 | |
| D9 | FJ907982 | 676(63.8) | 246(65.0) | 161(55.9) | V | -16.39 |
| D11 | FJ908056 | 679(50.5) | 242(50.0) | 161(42.9) | -9.42 | |
| D12 | FJ907981 | 677(64.0) | 246(65.0) | 161(55.3) | V | -16.90 |
| D13 | FJ907983 | 674(62.7) | 246(62.6) | 161(54.7) | V | -19.39 |
| D14 | FJ908045 | 658(50.2) | 243(50.6) | 161(43.5) | -10.37 | |
| D15 | FJ908067 | 671(48.7) | 243(50.2) | 161(44.7) | V | -13.21 |
| C1 | FJ907976 | 678(64.2) | 246(65.0) | 161(55.9) | V | -16.39 |
| C2 | FJ907979 | 678 (64.0) | 246(65.0) | 161(55.9) | V | -16.39 |
| C3 | FJ907978 | 679(64.1) | 246(65.0) | 161(55.3) | V | -16.59 |
| C4 | FJ907977 | 677 (64.1) | 246(64.6) | 161(55.9) | V | -16.39 |
| C5 | FJ907975 | 681(64.2) | 230(65.6) | 161(55.9) | V | -16.39 |
| ITS区域 ITS region | 序列类型 Sequence type | 序列数目 Sequence number | 多态位点数Polymorphic site no. | 总突变数 Total mutation | 核苷酸多态性 Nucleotide diversity (π) |
|---|---|---|---|---|---|
| ITS1 | cDNA ITS | 5 | 11 | 11 | 0.007 |
| 功能 ITS Functional ITS | 7 | 25 | 25 | 0.011 | |
| 假基因 Pseudogene | 7 | 258 | 283 | 0.191 | |
| ITS2 | cDNA ITS | 5 | 2 | 2 | 0.003 |
| 功能 ITS Functional ITS | 7 | 11 | 11 | 0.013 | |
| 假基因 Pseudogene | 7 | 120 | 142 | 0.222 | |
| 5.8S | cDNA ITS | 5 | 1 | 1 | 0.002 |
| 功能 ITS Functional ITS | 7 | 4 | 4 | 0.007 | |
| 假基因 Pseudogene | 7 | 64 | 72 | 0.174 | |
| ITS | cDNA ITS | 5 | 14 | 14 | 0.005 |
| 功能 ITS Functional ITS | 7 | 40 | 40 | 0.011 | |
| 假基因 Pseudogene | 7 | 442 | 497 | 0.196 |
表2 苏铁ITS核苷酸多态性
Table 2 Nucleotide diversity of the ITS region in Cycas revoluta
| ITS区域 ITS region | 序列类型 Sequence type | 序列数目 Sequence number | 多态位点数Polymorphic site no. | 总突变数 Total mutation | 核苷酸多态性 Nucleotide diversity (π) |
|---|---|---|---|---|---|
| ITS1 | cDNA ITS | 5 | 11 | 11 | 0.007 |
| 功能 ITS Functional ITS | 7 | 25 | 25 | 0.011 | |
| 假基因 Pseudogene | 7 | 258 | 283 | 0.191 | |
| ITS2 | cDNA ITS | 5 | 2 | 2 | 0.003 |
| 功能 ITS Functional ITS | 7 | 11 | 11 | 0.013 | |
| 假基因 Pseudogene | 7 | 120 | 142 | 0.222 | |
| 5.8S | cDNA ITS | 5 | 1 | 1 | 0.002 |
| 功能 ITS Functional ITS | 7 | 4 | 4 | 0.007 | |
| 假基因 Pseudogene | 7 | 64 | 72 | 0.174 | |
| ITS | cDNA ITS | 5 | 14 | 14 | 0.005 |
| 功能 ITS Functional ITS | 7 | 40 | 40 | 0.011 | |
| 假基因 Pseudogene | 7 | 442 | 497 | 0.196 |
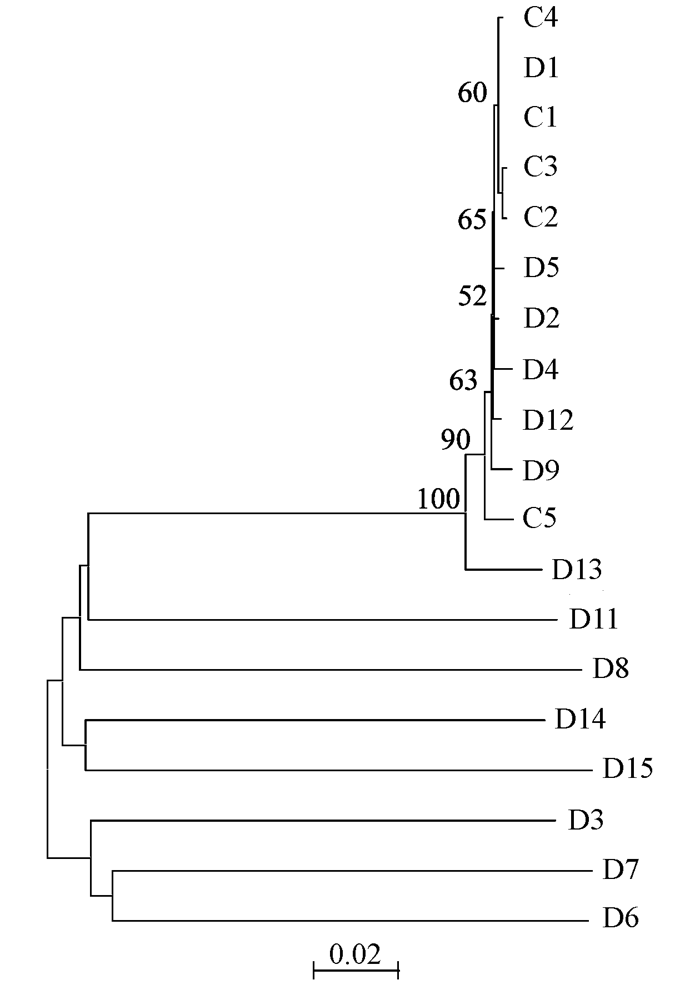
图1 基于Kimura双参数模型用苏铁检测到的所有ITS拷贝序列构建的NJ树。数字表示2,000次重复取样后大于50%的自展支持率。
Fig. 1 Neighbor-joining tree topology constructed on Kimura two-parameter distance matrix using ITS entire region of all paralogs in C. revoluta. Numbers indicate the over 50% bootstrap values with 2,000 replicates.
| [1] |
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution, 29,417-434.
DOI URL PMID |
| [2] |
Baker WJ, Hedderson TA, Dransfield J (2000) Molecular phylogenetics of subfamily Calamoideae (Palmae) based on nrDNA ITS and cpDNA rps16 intron sequence data. Molecular Phylogenetics and Evolution, 14,195-217.
URL PMID |
| [3] | Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA―a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden, 82,247-277. |
| [4] |
Buckler ES, Holtsford TP (1996) Zea ribosomal repeat evolution and mutation patterns. Molecular Biology and Evolution, 13,623-632.
DOI URL PMID |
| [5] |
Buckler ES, Ippolito A, Holtsford TP (1997) The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics, 145,821-832.
URL PMID |
| [6] |
Campbell CS, Wright WA, Cox M, Vining TF, Major CS, Arsenault MP (2005) Nuclear ribosomal DNA internal transcribed spacer 1 (ITS1) in Picea (Pinaceae), sequence divergence and structure. Molecular Phylogenetics and Evolution, 35,165-185.
URL PMID |
| [7] |
Chaw SM, Walters TW, Chang CC, Hu SH, Chen SH (2005) A phylogeny of cycads (Cycadales) inferred from chloroplast matk gene, trnK intron, and nuclear rDNA ITS region. Molecular Phylogenetics and Evolution, 37,214-234.
DOI URL PMID |
| [8] | Doyle J (1991) DNA protocols for plants - CTAB total DNA isolation. In: Molecular Techniques in Taxonomy (eds Hewitt GM, Johnston A), pp.283-293. Springer, Berlin. |
| [9] |
Eickbush TH, Eickbush DG (2007) Finely orchestrated movements, evolution of the ribosomal RNA genes. Genetics, 175,477-485.
DOI URL PMID |
| [10] | Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41,95-98. |
| [11] |
Harpke D, Peterson A (2006) Non-concerted ITS evolution in Mammillaria (Cactaceae). Molecular Phylogenetics and Evolution, 41,579-593.
DOI URL PMID |
| [12] | Hizume M, Ishida F, Kondo K (1992) Differential staining and in situ hybridization of nucleolar oraganizers and centromeres in Cycas revoluta chromosomes. Japanese Journal of Genetics, 67,381-387. |
| [13] | Jobes DV, Thien LB (1997) A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. Plant Molecular Biology Reporter, 15,326-334. |
| [14] | Johnson LAS, Wilson KL (1990) General traits of the Cycadales. In: The Families and Genera of Vascular Plants (eds Kramer KU, Green PS), pp.363-368. Springer-Verlag, Berlin. |
| [15] |
Kan XZ, Wang SS, Ding X, Wang XQ (2007) Structural evolution of nrDNA ITS in Pinaceae and its phylogenetic implications. Molecular Phylogenetics and Evolution, 44,765-777.
DOI URL PMID |
| [16] | Karvonen P, Savolainen O (1993) Variation and inheritance of ribosomal DNA in Pinus sylvestris L. (Scots pine). Heredity, 71,614-622. |
| [17] | Kumar S, Tamura K, Nei M (1994) MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Bioinformatics, 10,189-191. |
| [18] | Liston A, Robinson WA, Oliphant JM, Alvarez-Buylla ER (1996) Length variation in the nuclear ribosomal DNA internal transcribed spacer region of non-flowering seed plants. Systematic Botany, 21,109-120. |
| [19] |
Mayol M, Rossello JA (2001) Why nuclear ribosomal DNA spacers (ITS) tell different stories in Quercus. Molecular Phylogenetics and Evolution, 19,167-176.
DOI URL PMID |
| [20] |
Muir G, Fleming CC, Schlotterer C (2001) Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Molecular Biology and Evolution, 18,112-119.
URL PMID |
| [21] |
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annual Review of Genetics, 39,121-152.
DOI URL PMID |
| [22] | Nickrent DL, Schuette KP, Starr EM (1994) A molecular phylogeny of Arceuthbium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany, 81,1149-1160. |
| [23] | Osborne R (1995) The world cycad census and a proposed revision of the threatened species status for cycad taxa. Biological Conservation, 71,1-12. |
| [24] |
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology, 265,218-225.
DOI URL PMID |
| [25] |
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics, 15,174-175.
URL PMID |
| [26] | Sang T, Crawford J, Stuessy TF (1995) Documentation of reticulate evolution in peonies ( Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA, implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences, USA, 92,6813-6817. |
| [27] | Shimabuku K (1997) Check List Vascular Flora of the Ryukyu Islands, pp.1-855. Kyushu University Press, Fukuoka. (in Japanese). |
| [28] | Stevenson DW (1990) Morphology and systematics of the Cycadales. Memoirs of the New York Botanical Garden, 57,8-55. |
| [29] |
Suh Y, Thien LB, Zimmer EA (1992) Nucleotide sequences of the internal transcribed spacers and 5.8S rRNA gene in Canella winterana (Magnoliales; Canellaceae). Nucleic Acids Research, 20,6101-6102.
DOI URL PMID |
| [30] | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 24,4876-4882. |
| [31] |
Wei XX, Wang XQ (2004) Recolonization and radiation in Larix (Pinaceae): evidence from nuclear ribosomal DNA paralogues. Molecular Ecology, 13,3115-3123.
DOI URL PMID |
| [32] |
Wei XX, Wang XQ, Hong DY (2003) Marked intragenomic heterogeneity and geographical differentiation of nrDNA ITS in Larix potaninii (Pinaceae). Journal of Molecular Evolution, 57,623-635.
URL PMID |
| [33] | Whitelock LM (2002) The Cycads. Timber Press, Inc.,Portland. |
| [34] |
Won H, Renner SS (2005) The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Molecular Phylogenetics and Evolution, 36,581-597.
DOI URL PMID |
| [35] |
Zheng XY, Cai DY, Yao LH, Teng YW (2008) Non-concerted ITS evolution, early origin and phylogenetic utility of ITS pseudogenes in Pyrus. Molecular Phylogenetics and Evolution, 48,892-903.
URL PMID |
| [36] |
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31,3406-3415.
DOI URL PMID |
| [1] | 蔡新宇, 毛晓伟, 赵毅强. 家养动物驯化起源的研究方法与进展[J]. 生物多样性, 2022, 30(4): 21457-. |
| [2] | 莫日根高娃, 商辉, 刘保东, 康明, 严岳鸿. 一个种还是多个种? 简化基因组及其形态学证据揭示中国白桫椤植物的物种多样性分化[J]. 生物多样性, 2019, 27(11): 1196-1204. |
| [3] | 马洪峥, 李珊珊, 葛颂, 戴思兰, 陈文俐. 能源作物芒属双药芒组SSR引物的筛选及其评价[J]. 生物多样性, 2011, 19(5): 535-542. |
| [4] | 戴李川, 张明龙, 刘继业, 李小白, 崔海瑞. 用EST-SSR标记检测我国甘蓝型油菜品种的遗传多样性[J]. 生物多样性, 2009, 17(5): 482-489. |
| [5] | 潘兆娥, 孙君灵, 王西文, 贾银华, 周忠丽, 庞保印, 杜雄明. 基于棉花参比种质的SSR多态性核心引物筛选[J]. 生物多样性, 2008, 16(6): 555-561. |
| [6] | 董志军, 黄晖, 黄良民, 李元超, 李秀保. 运用PCR-RFLP方法研究三亚鹿回头岸礁造礁石珊瑚共生藻的组成[J]. 生物多样性, 2008, 16(5): 498-502. |
| [7] | 饶友生, 张细权. 拷贝数目变异: 基因组遗传变异的新诠释[J]. 生物多样性, 2008, 16(4): 399-406. |
| [8] | 张田, 李作洲, 刘亚令, 姜正旺, 黄宏文. 猕猴桃属植物的cpSSR遗传多样性及其同域分布物种的杂交渐渗与同塑[J]. 生物多样性, 2007, 15(1): 1-22. |
| [9] | 黄勃, 王成树, 王滨, 樊美珍, 李增智. 粉拟青霉种内nrDNA ITS分析[J]. 生物多样性, 2003, 11(6): 480-485. |
| [10] | 邹喻苹, 葛颂. 新一代分子标记——SNPs及其应用[J]. 生物多样性, 2003, 11(5): 370-382. |
| [11] | 赵桂仿, Francois Felber, Philippe Kuepfer. 阿尔卑斯山黄花茅(Anthoxanthum alpinum)沿海拔梯度的基因频率分布[J]. 生物多样性, 2000, 08(1): 95-102. |
| [12] | 虞泓, 郑树松, 黄瑞复. 云南松居群内雄球花多态性[J]. 生物多样性, 1998, 06(4): 267-271. |
| [13] | 冯书堂, 陈幼春. 五指山猪生物学特性、易地繁育及遗传多样性研究[J]. 生物多样性, 1998, 06(3): 172-179. |
| [14] | 吴平, 周开亚, 王亚明, 黄恭情, 徐麟木. 用RFLP和PCR-RFLP技术研究东北虎和华南虎线粒体DNA多态性*[J]. 生物多样性, 1997, 05(3): 173-178. |
| [15] | 于长青. 中国麋鹿遗传多样性现状与保护对策[J]. 生物多样性, 1996, 04(3): 130-134. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn