



Biodiv Sci ›› 2016, Vol. 24 ›› Issue (6): 694-700. DOI: 10.17520/biods.2016003 cstr: 32101.14.biods.2016003
Special Issue: 青藏高原生物多样性与生态安全
• Methodologies • Previous Articles Next Articles
Xiao Luo1,2, Feng Li1,3, Jing Chen1,2, Zhigang Jiang1,2,*( )
)
Received:2016-01-03
Accepted:2016-05-13
Online:2016-06-20
Published:2016-06-20
Contact:
Jiang Zhigang
Xiao Luo, Feng Li, Jing Chen, Zhigang Jiang. The taxonomic status of badgers in the Qinghai Lake area and evolutionary history of Meles[J]. Biodiv Sci, 2016, 24(6): 694-700.
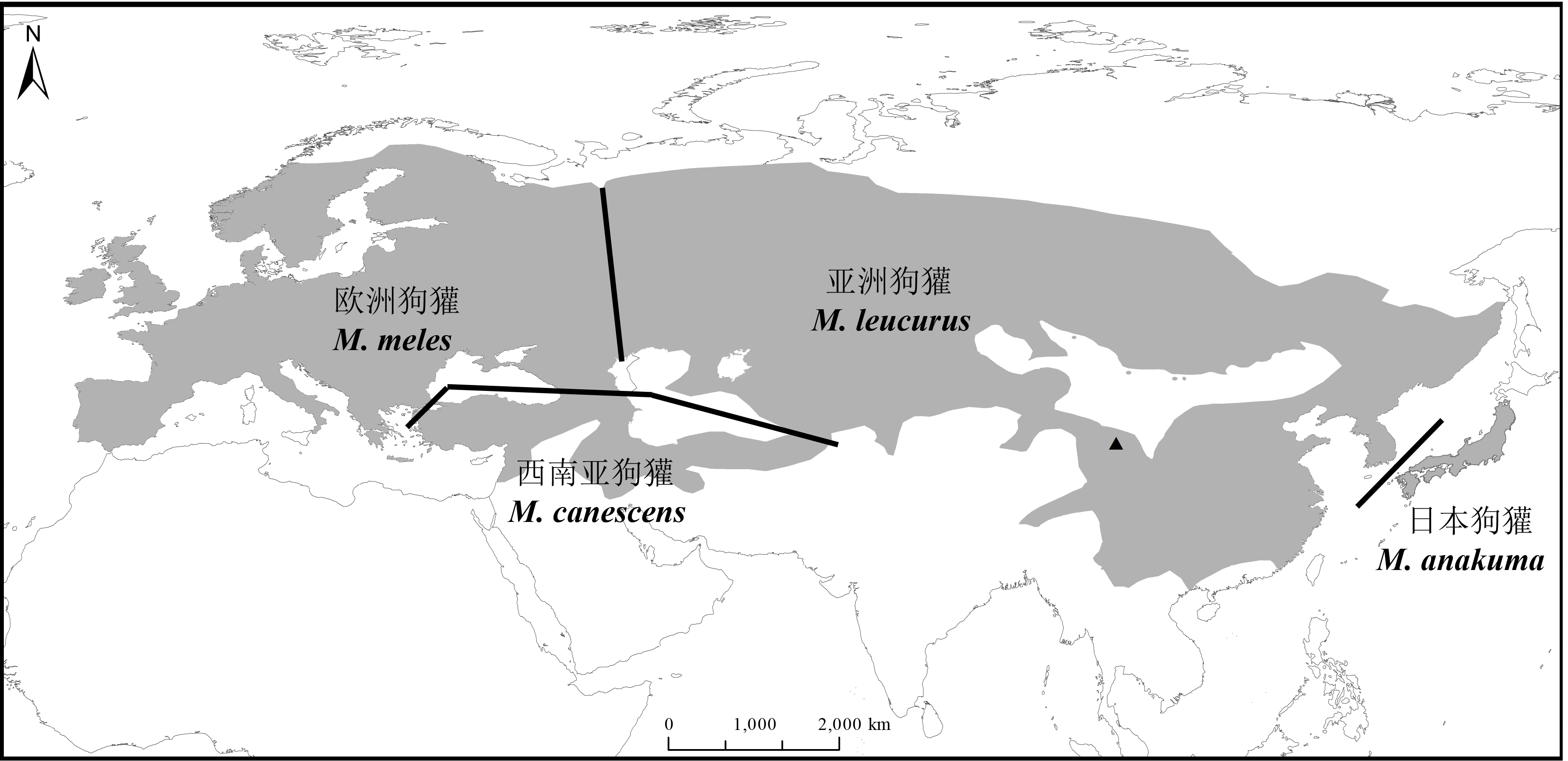
Fig. 1 Geographic distribution of Eurasian badger. The grey area shows the range of Eurasian badger, the black lines represent geographic boundaries of each phylogenetic group and the black triangle indicates our sampling locality.
| 单倍型 Haplotype | 样本代码 Sample code | 样本编号 Sample no. | 采样点 Sampling locality | 序列号 Accession no. | |
|---|---|---|---|---|---|
| Cyt b | Control region | ||||
| H1 | QH1 | B1 | 中国青海 Qinghai, China | KU361236 | KU361238 |
| H2 | QH2 | B2 | 中国青海 Qinghai, China | KU361237 | KU361239 |
| H2 | QH3 | B3 | 中国青海 Qinghai, China | KU361237 | KU361239 |
| H3 | QH4 | B4 | 中国青海 Qinghai, China | - | KU361239 |
| H4 | QH5 | B5 | 中国青海 Qinghai, China | - | KU361240 |
| H4 | QH6 | B6 | 中国青海 Qinghai, China | - | KU361240 |
| H5 | China | - | 中国黑龙江 Heilongjiang, China | KU052604 | KU052604 |
| H6 | Sweden | - | 瑞典 Sweden | AM711900 | AM711900 |
| H7 | Mongolia | 45Mo | 蒙古 Mongolia | HQ711950 | AJ563694 |
| H8 | Greece | 80Cr | 希腊克里特岛 Crete Island, Greece | HQ711947 | GU247573 |
| H9 | Israel1 | 49Is | 以色列 Israel | HQ711946 | AJ563686 |
| H10 | Israel2 | 48Is | 以色列 Israel | HQ711945 | AJ563685 |
| H11 | Spain | 35Sp | 西班牙 Spain | HQ711943 | AJ563676 |
| H12 | Russia1 | ZIS33 | 俄罗斯外贝加尔地区 Transbaikalia, Russia | AB049807 | AB538995 |
| H13 | Russia2 | ZIS36 | 俄罗斯列宁格勒州 Leningrad Province, Russia | AB049808 | AB538997 |
| H14 | Russia3 | ZIS35 | 俄罗斯列宁格勒州 Leningrad Province, Russia | AB049809 | AB538998 |
| H15 | Japan1 | K1 | 日本九州大分县 Oita, Kyushu, Japan | AB049806 | AB538971 |
| H16 | Japan2 | K6 | 日本九州大分县 Oita, Kyushu, Japan | AB049800 | AB538971 |
| H17 | Japan3 | K7 | 日本九州大分县 Oita, Kyushu, Japan | AB049802 | AB538971 |
| H18 | Japan4 | K8 | 日本九州福冈县 Fukuoka, Kyushu, Japan | AB049799 | AB538971 |
| H19 | Japan5 | YMG1 | 日本本州山口县 Yamaguchi, Honshu, Japan | AB049795 | AB538983 |
| H20 | Japan6 | MR1 | 日本九州岩手县 Iwate, Kyushu, Japan | AB049791 | AB538977 |
| H21 | Japan7 | KPM-NF1002945 | 日本本州神奈川县 Kanagawa, Honshu, Japan | AB291075 | AB291075 |
| Arctonyx collaris | YP6001 | HM106329 | HM106329 | ||
Table 1 The mitochondrial DNA sequences information in this study
| 单倍型 Haplotype | 样本代码 Sample code | 样本编号 Sample no. | 采样点 Sampling locality | 序列号 Accession no. | |
|---|---|---|---|---|---|
| Cyt b | Control region | ||||
| H1 | QH1 | B1 | 中国青海 Qinghai, China | KU361236 | KU361238 |
| H2 | QH2 | B2 | 中国青海 Qinghai, China | KU361237 | KU361239 |
| H2 | QH3 | B3 | 中国青海 Qinghai, China | KU361237 | KU361239 |
| H3 | QH4 | B4 | 中国青海 Qinghai, China | - | KU361239 |
| H4 | QH5 | B5 | 中国青海 Qinghai, China | - | KU361240 |
| H4 | QH6 | B6 | 中国青海 Qinghai, China | - | KU361240 |
| H5 | China | - | 中国黑龙江 Heilongjiang, China | KU052604 | KU052604 |
| H6 | Sweden | - | 瑞典 Sweden | AM711900 | AM711900 |
| H7 | Mongolia | 45Mo | 蒙古 Mongolia | HQ711950 | AJ563694 |
| H8 | Greece | 80Cr | 希腊克里特岛 Crete Island, Greece | HQ711947 | GU247573 |
| H9 | Israel1 | 49Is | 以色列 Israel | HQ711946 | AJ563686 |
| H10 | Israel2 | 48Is | 以色列 Israel | HQ711945 | AJ563685 |
| H11 | Spain | 35Sp | 西班牙 Spain | HQ711943 | AJ563676 |
| H12 | Russia1 | ZIS33 | 俄罗斯外贝加尔地区 Transbaikalia, Russia | AB049807 | AB538995 |
| H13 | Russia2 | ZIS36 | 俄罗斯列宁格勒州 Leningrad Province, Russia | AB049808 | AB538997 |
| H14 | Russia3 | ZIS35 | 俄罗斯列宁格勒州 Leningrad Province, Russia | AB049809 | AB538998 |
| H15 | Japan1 | K1 | 日本九州大分县 Oita, Kyushu, Japan | AB049806 | AB538971 |
| H16 | Japan2 | K6 | 日本九州大分县 Oita, Kyushu, Japan | AB049800 | AB538971 |
| H17 | Japan3 | K7 | 日本九州大分县 Oita, Kyushu, Japan | AB049802 | AB538971 |
| H18 | Japan4 | K8 | 日本九州福冈县 Fukuoka, Kyushu, Japan | AB049799 | AB538971 |
| H19 | Japan5 | YMG1 | 日本本州山口县 Yamaguchi, Honshu, Japan | AB049795 | AB538983 |
| H20 | Japan6 | MR1 | 日本九州岩手县 Iwate, Kyushu, Japan | AB049791 | AB538977 |
| H21 | Japan7 | KPM-NF1002945 | 日本本州神奈川县 Kanagawa, Honshu, Japan | AB291075 | AB291075 |
| Arctonyx collaris | YP6001 | HM106329 | HM106329 | ||
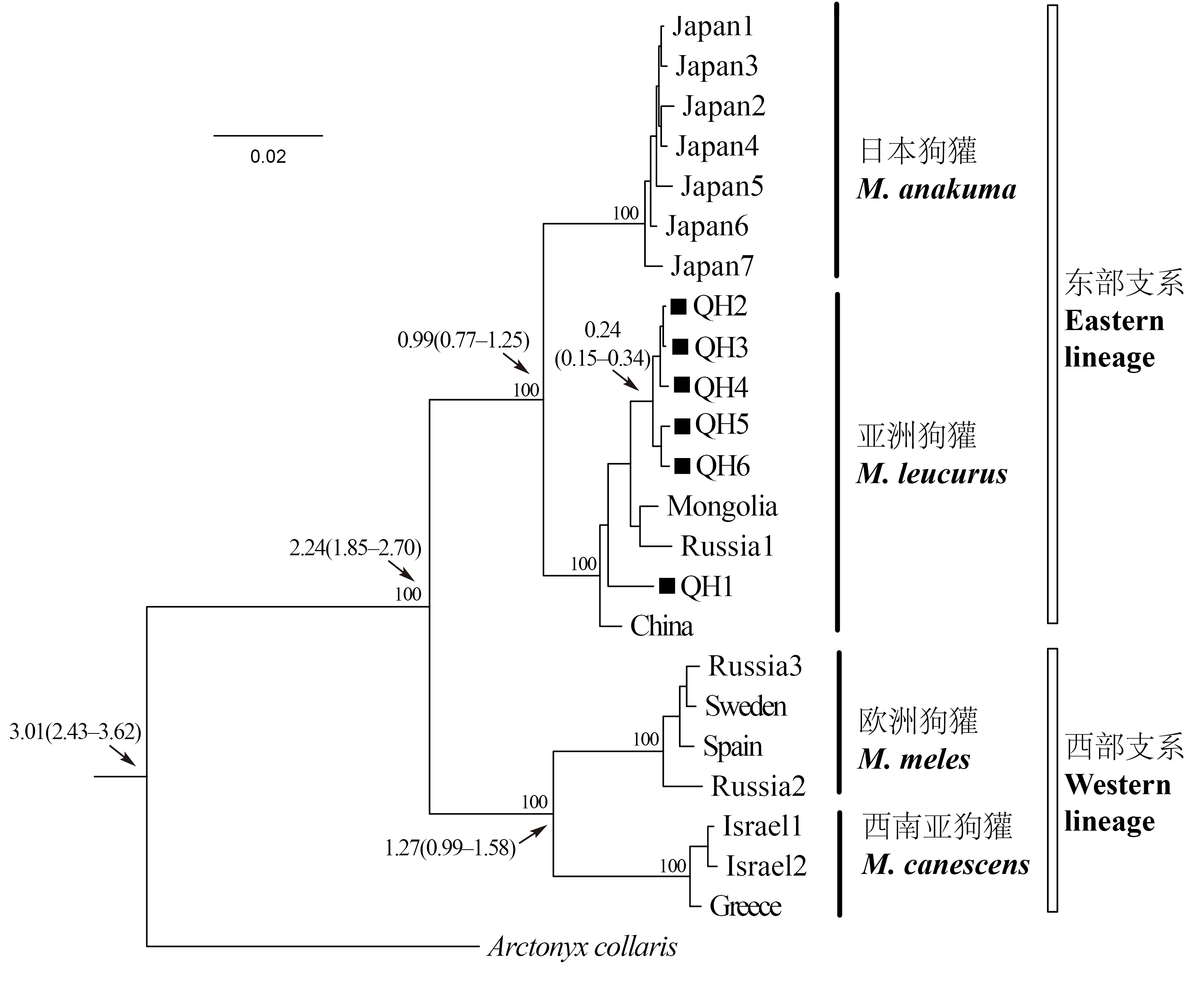
Fig. 2 Phylogenetic tree of mitochondrial DNA obtained from Bayesian analysis. The sample codes correspond with those in Table 1, in which QH1-6 represent badgers of Qinghai Lake area. The numbers on the branch indicate Bayesian posterior probability and estimated divergence time (with 95% HPD).
| 西南亚狗獾 M. canescens | 欧洲狗獾 M. meles | 日本狗獾 M. anakuma | 亚洲狗獾 M. leucurus | |
|---|---|---|---|---|
| 欧洲狗獾 M. meles | 0.034 ± 0.007 | |||
| 日本狗獾 M. anakuma | 0.042 ± 0.009 | 0.043 ± 0.008 | ||
| 亚洲狗獾 M. leucurus | 0.040 ± 0.008 | 0.041 ± 0.008 | 0.021 ± 0.005 | |
| 青海湖地区狗獾 Badger in the Qinghai Lake area | 0.038 ± 0.008 | 0.041 ± 0.008 | 0.020 ± 0.006 | 0.017 ± 0.004 |
Table 2 Genetic distances among different Eurasian badger groups
| 西南亚狗獾 M. canescens | 欧洲狗獾 M. meles | 日本狗獾 M. anakuma | 亚洲狗獾 M. leucurus | |
|---|---|---|---|---|
| 欧洲狗獾 M. meles | 0.034 ± 0.007 | |||
| 日本狗獾 M. anakuma | 0.042 ± 0.009 | 0.043 ± 0.008 | ||
| 亚洲狗獾 M. leucurus | 0.040 ± 0.008 | 0.041 ± 0.008 | 0.021 ± 0.005 | |
| 青海湖地区狗獾 Badger in the Qinghai Lake area | 0.038 ± 0.008 | 0.041 ± 0.008 | 0.020 ± 0.006 | 0.017 ± 0.004 |
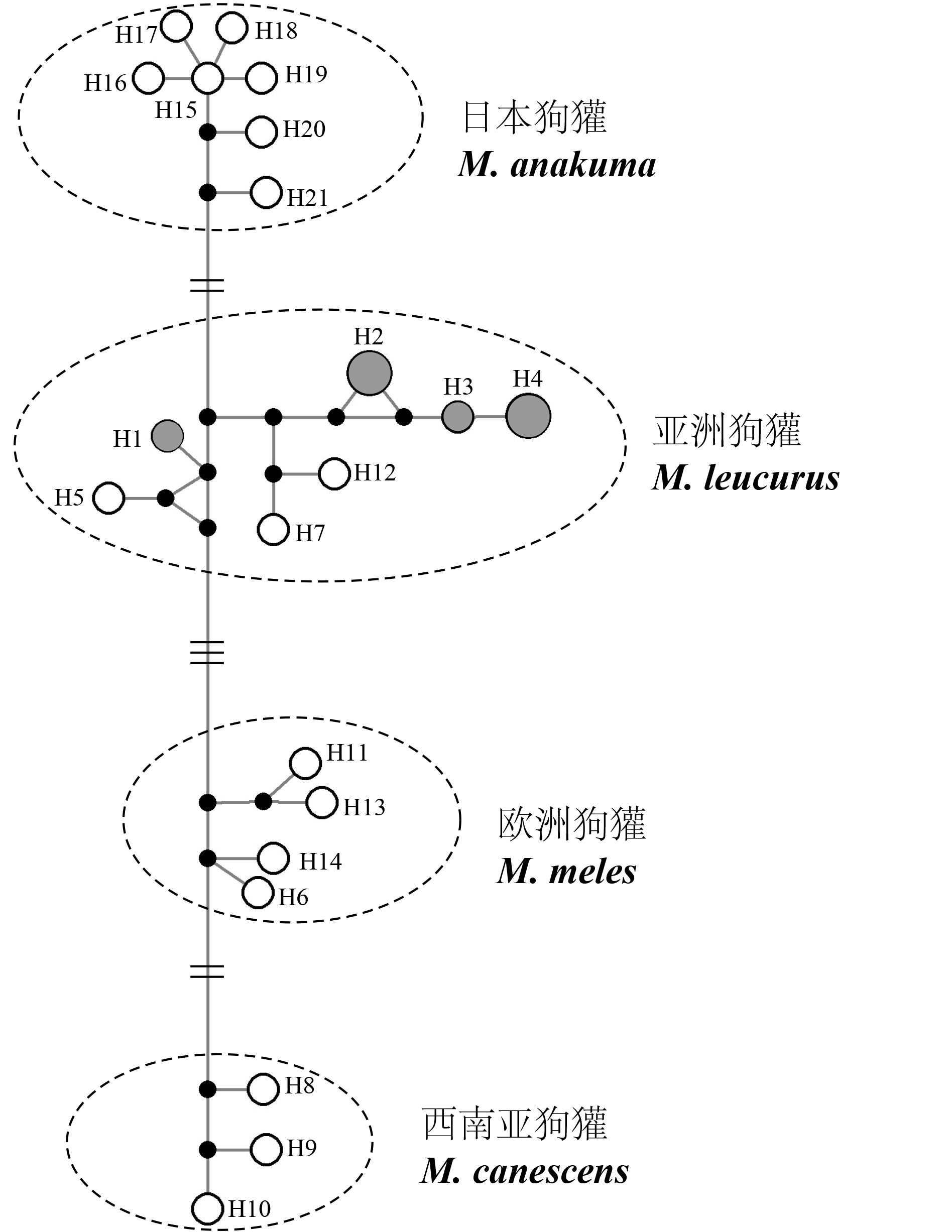
Fig. 3 The network based on 21 haplotypes of Eurasian badgers mitochondrial DNA. H1-4 represent Qinghai badgers (in grey). All haplotypes are displayed in Table 1. The black circles represent missing haplotypes. The sizes of the white circles in the network represent the frequencies of each haplotype. Four badger’s groups are circled by dotted line.
| [1] | Abramov AV (2001) Notes on the taxonomy of the Siberian badgers (Mustelidae: Meles). Proceedings of the Zoological Institute, Russian Academy of Sciences, 288, 221-233. |
| [2] | Abramov AV (2002) Variation of the baculum structure of the Palaearctic badger (Carnivora, Mustelidae, Meles). Russian Journal of Theriology, 1, 57-60. |
| [3] | Abramov AV, Puzachenko AY (2005) Sexual dimorphism of craniological characters in Eurasian badgers, Meles spp. (Carnivora, Mustelidae). Zoologischer Anzeiger, 244, 11-29. |
| [4] | Abramov AV, Puzachenko AY (2006) Geographical variability of skull and taxonomy of Eurasian badgers (Mustelidae, Meles). Zoologicheskii Zhurnal, 85, 641-655. (in Russian with English abstract) |
| [5] | Abramov AV, Puzachenko AY (2013) The taxonomic status of badgers (Mammalia, Mustelidae) from Southwest Asia based on cranial morphometrics, with the redescription of Meles canescens. Zootaxa, 3681(1), 044-058. |
| [6] | Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| [7] | Baryshnikov GF, Potapova OR (1990) Variability of the dental system in badgers (Meles, Carnivora) of the USSR fauna. Zoologicheskii Zhurnal, 69, 84-97. (in Russian with English abstract) |
| [8] | Corbet GB (1978) The Mammals of the Palaearctic Region: A Taxonomic Review. Cornell University Press, London and Ithaca. |
| [9] | Del Cerro I, Marmi J, Ferrando A, Chashchin P, Taberlet P, Bosch M (2010) Nuclear and mitochondrial phylogenies provide evidence for four species of Eurasian badgers (Carnivora). Zoologica Scripta, 39, 415-425. |
| [10] | Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214. |
| [11] | Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969-1973. |
| [12] | Frantz AC, McDevitt AD, Pope LC, Kochan J, Davison J, Clements CF, Elmeros M, Molina-Vacas G, Ruiz-Gonzalez A, Balestrieri A, Berge KVD, Breyne P, Do Linh San E, Gren E, Suchentrunk F, Schley L, Kowalczyk R, Kostka BI, Cirovic D, Sprem N, Colyn M, Ghirardi M, Racheva V, Braun C, Oliveira R, Lanszki J, Stubbe A, Stubbe M, Stier N, Burke T (2014) Revisiting the phylogeography and demography of European badgers (Meles meles) based on broad sampling, multiple markers and simulations. Heredity, 113, 443-453. |
| [13] | Gao YT (1987) Fauna Sinica, Mammalia: Vol. 8, Carnivora, pp. 214-223. Science Press, Beijing. (in Chinese) |
| [高耀亭 (1987) 中国动物志·兽纲: 第八卷, 食肉目, 214-223页. 科学出版社, 北京.] | |
| [14] | Koepfli KP, Deere KA, Slater GJ, Begg C, Begg K, Grassman L, Lucherini M, Veron G, Wayne RK (2008) Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biology, 6, 1-22. |
| [15] | Koh HS, Kryukov A, Oh JG, Bayarkhagva D, Yang BG, Ahn NH, Bazarsad D (2014) Two sympatric phylogroups of the Asian badger Meles leucurus (Carnivora: Mammalia) identified by mitochondrial DNA cytochrome b gene sequences. Russian Journal of Theriology, 13, 1-8. |
| [16] | Kurose N, Abramov AV, Masuda R (2000) Intrageneric diversity of the cytochrome b gene and phylogeny of Eurasian species of the genus Mustela (Mustelidae, Carnivora). Zoological Science, 17, 673-679. |
| [17] | Kurten B (1968) The Pleistocene Mammals of Europe. Weidenfeld & Nicolsan, London. |
| [18] | Li F, Jiang ZG (2014) Is nocturnal rhythm of Asian badger (Meles leucurus) caused by human activity? A case study in the eastern area of Qinghai Lake. Biodiversity Science, 22, 758-763. (in Chinese with English abstract) |
| [李峰, 蒋志刚 (2014) 狗獾夜间活动节律是受人类活动影响而形成的吗? 基于青海湖地区的研究实例. 生物多样性, 22, 758-763.] | |
| [19] | Li F, Luo ZH, Li CL, Li CW, Jiang ZG (2013) Biogeographical patterns of the diet of Palearctic badger: Is badger an earthworm specialist predator? Chinese Science Bulletin, 58, 2255-2261. |
| [20] | Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452. |
| [21] | Lynch JM (1994) Morphometric variation in the badger (Meles meles): clinal variation in cranial size and shape across Eurasia. Small Carnivore Conservation, 10, 6-7. |
| [22] | Madurell-Malapeira J, Alba DM, Marmi J, Aurell J, Moyà-Solà S (2011) The taxonomic status of European Plio-Pleistocene badgers. Journal of Vertebrate Paleontology, 31, 885-894. |
| [23] | Marmi J, López-Giráldez F, MacDonald DW, Calafell E, Zholnerovskaya E, Domingo-Roura X (2006) Mitochondrial DNA reveals a strong phylogeographic structure in the badger across Eurasia. Molecular Ecology, 15, 1007-1020. |
| [24] | Marmi J, López-Giráldez JF, Domingo-Roura X (2004) Phylogeny, evolutionary history and taxonomy of the Mustelidae based on sequences of the cytochrome b gene and a complex repetitive flanking region. Zoologica Scripta, 33, 481-499. |
| [25] | Neal E, Cheeseman C (1996) Badgers. T & AD Poyser Ltd., London. |
| [26] | Nowak RM, Paradiso JL (1999) Walker’s Mammals of the World. Cambridge University Press, London. |
| [27] | Petter G (1971) Origine, phylogenie et systematique des blaireaux. Mammalia, 35, 567-597. (in French) |
| [28] | Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics, 14, 817-818. |
| [29] | Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574. |
| [30] | Song G, Qu Y, Yin Z, Li S, Liu N, Lei F (2009) Phylogeography of the Alcippe morrisonia (Aves: Timaliidae): long population history beyond late Pleistocene glaciations. BMC Evolutionary Biology, 9, 143. |
| [31] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725-2729. |
| [32] | Tashima S, Kaneko Y, Anezaki T, Baba M, Yachimori S, Abramov AV, Saveljev AP, Masuda R (2011a) Phylogeographic sympatry and isolation of the Eurasian badgers (Meles, Mustelidae, Carnivora): implications for an alternative analysis using maternally as well as paternally inherited genes. Zoological Science, 28, 293-303. |
| [33] | Tashima S, Kaneko Y, Anezaki T, Baba M, Yachimori S, Abramov AV, Saveljev AP, Masuda R (2011b) Identification and molecular variations of CAN-SINEs from the ZFY gene final intron of the Eurasian badgers (genus Meles). Mammal Study, 36, 41-48. |
| [34] | Thompson JD, Gibson T, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics, Chapter 2 (Unit 2.3), doi: 10.1002/0471250953.bi0203s00. |
| [35] | Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics, 27, 171-180. |
| [36] | Viret J (1950) Meles thorali n. sp. du loess villafranchien de Saint-Vallier (Drôme). Eclogae Geologicae Helvetiae, 43, 274-287. (in French with English abstract) |
| [37] | Wozencraft WC (1993) Order Carnivora. In: Mammal Species of the World: A Taxonomic and Geographic Reference, 2nd edn. (eds Wilson DE, Reeder DM), pp. 279-348. Smithsonian Institution Press, Washington, DC. |
| [38] | Wozencraft WC (2005) Order Carnivora. In: Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd edn. (eds Wilson DE, Reeder DM), pp. 532-628. Johns Hopkins University Press, Baltimore. |
| [39] | Xie ZG (2011) Studies on the Population Ecology of Badger (Meles meles) in Shanghai. PhD dissertation, East China Normal University, Shanghai. (in Chinese with English abstract) |
| [谢志刚 (2011) 上海地区狗獾生态学研究. 博士学位论文, 华东师范大学, 上海.] | |
| [40] | Xu X, Xie ZG, Cui YY, Chu KL, Jiang WZ, Pei EL, Xu HF (2012) Activity patterns of reintroduced badgers in seminatural area. Chinese Journal of Zoology, 47(3), 49-52. (in Chinese with English abstract) |
| [徐循, 谢志刚, 崔勇勇, 褚可龙, 蒋文忠, 裴恩乐, 徐宏发 (2012) 重引入狗獾秋冬季行为的初步研究. 动物学杂志, 47(3), 49-52.] | |
| [41] | Yang HT, Liu ZS, Xu K, Song CL, Wu MF, Sun JH (2010) Autumn habitat selection of Eurasian badgers (Meles meles amurensis): a case of Fangzheng Forestry Bureau, Heilongjiang Province, China. Acta Ecologica Sinica, 30, 1875-1881. (in Chinese with English abstract) |
| [杨会涛, 刘振生, 徐坤, 宋丛亮, 吴木芬, 孙景海 (2010) 狗獾秋季对生境的选择——以黑龙江省方正林业局为例. 生态学报, 30, 1875-1881.] | |
| [42] | Ye XD, Ma Y, Wang RH, Dong AY (2000) Review of the diet of Eurasian badgers. Chinese Journal of Zoology, 35(2), 43-50. (in Chinese) |
| [叶晓堤, 马勇, 王润海, 董安渝 (2000) 欧亚大陆狗獾食性的研究概述. 动物学杂志, 35(2), 43-50.] | |
| [43] | Zhou WW, Yu L, Tan BY, Liu YT, Zhang L, Hua Y (2015) Phylogenetic relationship of Asian badger Meles leucurus amurensis revealed by complete mitochondrial genome. Mitochondrial DNA, published online, doi: 10.3109/ 19401736.2015.1127365. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()