



Biodiv Sci ›› 2015, Vol. 23 ›› Issue (4): 550-555. DOI: 10.17520/biods.2015120 cstr: 32101.14.biods.2015120
• Software introduction • Previous Articles
Xiaoting Xu1, Zhiheng Wang1,*( ), Dimitar Dimitrov2, Carsten Rahbek3,4
), Dimitar Dimitrov2, Carsten Rahbek3,4
Received:2015-05-07
Accepted:2015-07-09
Online:2015-07-20
Published:2015-08-03
Contact:
Wang Zhiheng
Xiaoting Xu, Zhiheng Wang, Dimitar Dimitrov, Carsten Rahbek. Using NCBIminer to search and download nucleotide sequences from GenBank[J]. Biodiv Sci, 2015, 23(4): 550-555.
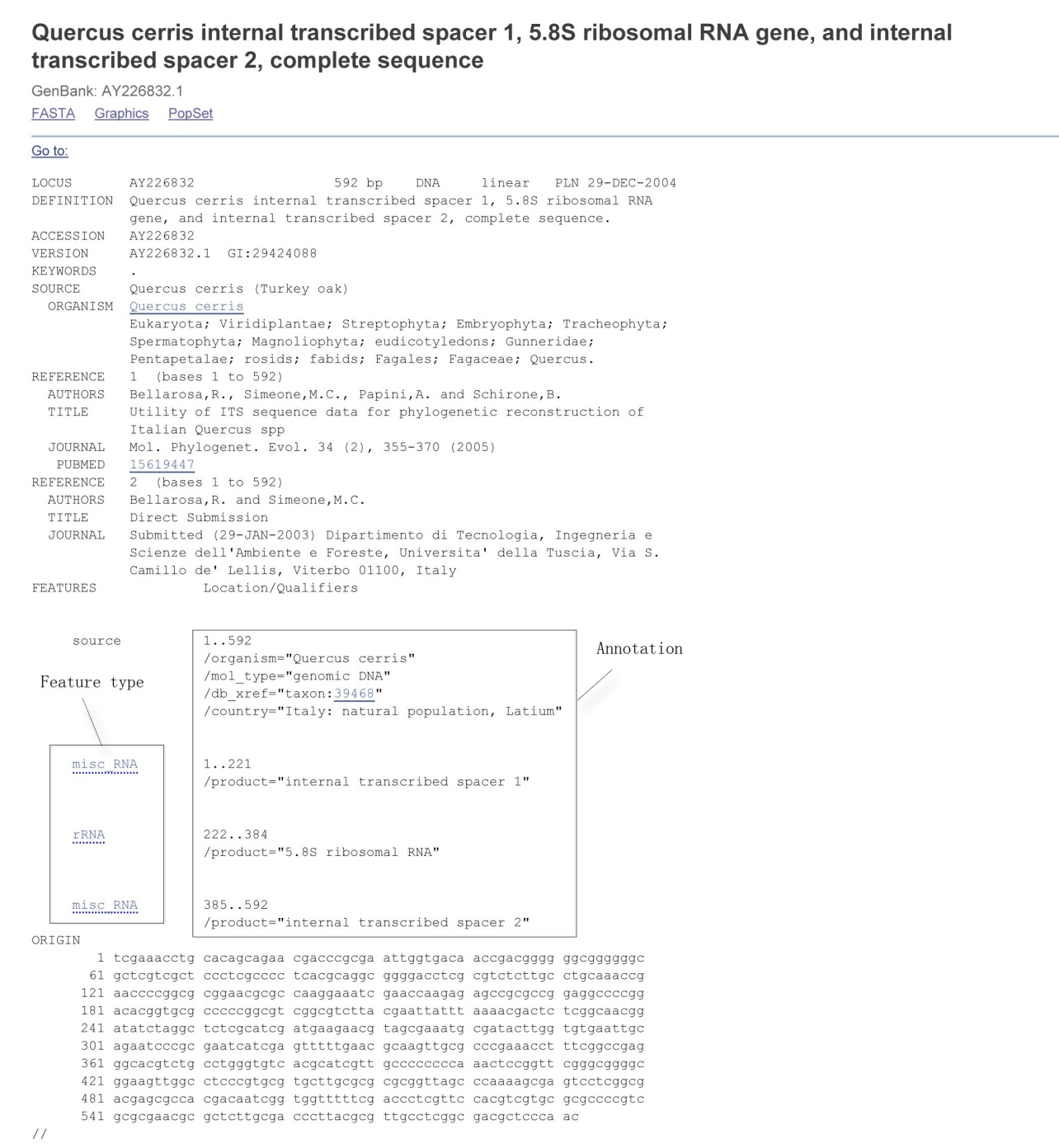
Appendix 1 Data format for a sequence in GenBank. The items in the left box are feature types defined in GenBank, while those in the right box are GenBank annotation information.
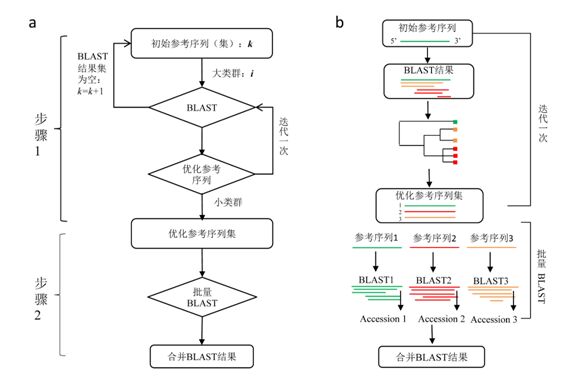
Appendix 2 Data format for a sequence in GenBank. The items in the left box are feature types defined in GenBank, while those in the right box are GenBank annotation informatioppendix 2 NCBIminer workflow. a, Major steps of the NCBIminer’s work flow; b, The algorithms for the establishment of improved reference sequences and sequence combination of multiple queries. Modified from Xu et al. (2015).
| 1 | Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool.Journal of Molecular Biology, 215, 403-410. |
| 2 | Chen ZD (陈之端), Li DZ (李德铢) (2013) On Barcode of Life and Tree of Life.Plant Diversity and Resources(植物分类与资源学报), 35, 675-681. (in Chinese with English abstract) |
| 3 | Driskell AC, Ané C, Burleigh JG, McMahon MM, O’Meara BC, Sanderson MJ (2004) Prospects for building the Tree of Life from large sequence databases.Science, 306, 1172-1174. |
| 4 | Holt B, Lessard JP, Borregaard MK, Fritz SA, Araujo MB, Dimitrov D, Fabre PH, Graham CH, Graves GR, Jonsson KA, Nogues-Bravo D, Wang ZH, Whittaker RJ, Fjeldsa J, Rahbek C (2013) An update of Wallace’s zoogeographic regions of the world.Science, 339, 74-78. |
| 5 | Jones M, Koutsovoulos G, Blaxter M (2011) iPhy: an integrated phylogenetic workbench for supermatrix analyses.BMC Bioinformatics, 12, 30. |
| 6 | Li DC (2013) Similarity analysis of DNA sequences based on CLZ complexity.Journal of Computational and Theoretical Nanoscience, 10, 481-487. |
| 7 | Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW, Grp CPB (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences, USA, 108, 19641-19646. |
| 8 | Lu LM (鲁丽敏), Sun M (孙苗), Zhang JB (张景博), Li HL (李洪雷), Lin L (林立), Yang T (杨拓), Chen M (陈闽), Chen ZD (陈之端) (2014) Tree of Life and its applications.Biodiversity Science(生物多样性), 22, 3-20. (in Chinese with English abstract) |
| 9 | Pearse WD, Purvis A (2013) phyloGenerator: an automated phylogeny generation tool for ecologists.Methods in Ecology and Evolution, 4, 692-698. |
| 10 | Pei NC (裴男才) (2015) Applications of DNA barcoding in evolutionary ecology.Biodiversity Science(生物多样性), 23, 291-292. (in Chinese) |
| 11 | Qiu Q, Zhang GJ, Ma T, Qian WB, Wang JY, Ye ZQ, Cao CC, Hu QJ, Kim J, Larkin DM, Auvil L, Capitanu B, Ma J, Lewin HA, Qian XJ, Lang YS, Zhou R, Wang LZ, Wang K, Xia JQ, Liao SG, Pan SK, Lu X, Hou HL, Wang Y, Zang XT, Yin Y, Ma H, Zhang J, Wang ZF, Zhang YM, Zhang DW, Yonezawa T, Hasegawa M, Zhong Y, Liu WB, Zhang Y, Huang ZY, Zhang SX, Long RJ, Yang HM, Wang J, Lenstra JA, Cooper DN, Wu Y, Wang J, Shi P, Wang J, Liu JQ (2012) The yak genome and adaptation to life at high altitude.Nature Genetics, 44, 946-949. |
| 12 | Ren BQ (任保青), Chen ZD (陈之端) (2010) DNA barcoding plant life.Chinese Bulletin of Botany(植物学报), 45, 1-12. (in Chinese with English abstract) |
| 13 | Sanderson M, Boss D, Chen D, Cranston K, Wehe A (2008) The PhyLoTA browser: processing GenBank for molecular phylogenetics research.Systematic Biology, 57, 335-346. |
| 14 | Xu X, Wang Z, Rahbek C, Lessard J-P, Fang J (2013) |
| 15 | Evolutionary history influences the effects of water-energy dynamics on oak diversity in Asia.Journal of Biogeography, 40, 2146-2155. |
| 16 | Xu XT, Dimitrov D, Rahbek C, Wang ZH (2015) NCBIminer: sequences harvest from Genbank.Ecography, 38, 426-430. |
| 17 | Yang ZY, Ran JH, Wang XQ (2012) Three genome-based phylogeny of Cupressaceae s.l.: further evidence for the evolution of gymnosperms and southern hemisphere biogeography.Molecular Phylogenetics and Evolution, 64, 452-470. |
| 18 | Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, Royer DL, Soltis DE, Stevens PF, Westoby M, Wright IJ, Aarssen L, Bertin RI, Calaminus A, Govaerts R, Hemmings F, Leishman MR, Oleksyn J, Soltis PS, Swenson NG, Warman L, Beaulieu JM (2013) Three keys to the radiation of angiosperms into freezing environments.Nature, 506, 89-92. |
| [1] | Tong Miao, Wang Huan, Zhang Wenshuang, Wang Chao, Song Jianxiao. Distribution characteristics of antibiotic resistance genes in soil bacterial communities exposed to heavy metal pollution [J]. Biodiv Sci, 2025, 33(3): 24101-. |
| [2] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [3] | Liu Zhixiang, Xie Hua, Zhang Hui, Huang Xiaolei. Functional diversity and regulation of cuticular hydrocarbons in social insects [J]. Biodiv Sci, 2025, 33(2): 24302-. |
| [4] | Cao Dong, Li Huanlong, Peng Yang, Wei Cunzheng. Progresses in the study of the relationship between plant genome size and traits [J]. Biodiv Sci, 2025, 33(2): 24192-. |
| [5] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [6] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [7] | Jiabei He, Ke Ke, Haiming Sun, Liping Hu, Xiaowei Zhao, Wenhao Wang, Qiang Zhao. Diet analysis of Neptunea cumingii using metabarcoding [J]. Biodiv Sci, 2025, 33(1): 24403-. |
| [8] | Wenjie Qu, Lei Wang, Wenyan Kang, Xinguo Yang, Jianjun Qu, Xue Zhang. Seed supply and regeneration potential of sand-fixing vegetation with different establishment years in the southeastern edge of the Tengger Desert [J]. Biodiv Sci, 2025, 33(1): 24254-. |
| [9] | Sicheng Han, Daowei Lu, Yuchen Han, Ruohan Li, Jing Yang, Ge Sun, Lu Yang, Junwei Qian, Xiang Fang, Shu-Jin Luo. Distribution of leopard cats in the nearest mountains to urban Beijing and its affecting environmental factors [J]. Biodiv Sci, 2024, 32(8): 24138-. |
| [10] | Yihui Jiang, Yue Liu, Xu Zeng, Zheying Lin, Nan Wang, Jihao Peng, Ling Cao, Cong Zeng. Fish diversity and connectivity in six national marine protected areas in the East China Sea [J]. Biodiv Sci, 2024, 32(6): 24128-. |
| [11] | Fuwei Zhao, Yingshuo Li, Hui Chen. Reflections on biodiversity legislation in China’s new era [J]. Biodiv Sci, 2024, 32(5): 24027-. |
| [12] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [13] | Yaoqi Chen, Jingjing Guo, Guojun Cai, Yili Ge, Yu Liao, Zheng Dong, Hui Fu. Evolution characteristics of submerged macrophyte community diversity in the middle and lower reaches of the Yangtze River in the past seventy years (1954-2021) [J]. Biodiv Sci, 2024, 32(3): 23319-. |
| [14] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [15] | Yang Ding, Yingqun Feng, Jinyu Zhang, Bo Wang. Seed predation and dispersal by animals of an endangered endemic species Pinus dabeshanensis [J]. Biodiv Sci, 2024, 32(3): 23401-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()