



Biodiv Sci ›› 2023, Vol. 31 ›› Issue (11): 23160. DOI: 10.17520/biods.2023160 cstr: 32101.14.biods.2023160
• Original Papers: Genetic Diversity • Next Articles
Yiyue He1, Yuying Liu1, Fubin Zhang3,4, Qiang Qin3,4, Yu Zeng1,4, Zhenyu Lü1, Kun Yang1,2,4,*( )
)
Received:2023-05-22
Accepted:2023-10-18
Online:2023-11-20
Published:2023-11-29
Contact:
* E-mail: About author:# Co-first authors
Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River[J]. Biodiv Sci, 2023, 31(11): 23160.
| 种群 Populations | 样本数(尾) Sample size | 体长 Body length (mm) | 湿重 Wet weight (g) | 纬度 Latitude | 经度 Longitude |
|---|---|---|---|---|---|
| 朝天区 CT | 27 | 100.12 ± 26.81 | 15.71 ± 10.27 | 32.65° N | 105.89° E |
| 苍溪县 CX | 28 | 132.04 ± 11.15 | 27.75 ± 6.47 | 31.74° N | 105.96° E |
| 蓬安县 PA | 30 | 109.95 ± 16.58 | 25.43 ± 33.82 | 31.04° N | 106.40° E |
| 合川区 HC | 28 | 114.84 ± 11.07 | 17.42 ± 4.40 | 29.98° N | 106.29° E |
Table 1 Sample information of four Saurogobio dabryi populations. CT, Chaotian; CX, Cangxi; PA, Peng’an; HC, Hechuan.
| 种群 Populations | 样本数(尾) Sample size | 体长 Body length (mm) | 湿重 Wet weight (g) | 纬度 Latitude | 经度 Longitude |
|---|---|---|---|---|---|
| 朝天区 CT | 27 | 100.12 ± 26.81 | 15.71 ± 10.27 | 32.65° N | 105.89° E |
| 苍溪县 CX | 28 | 132.04 ± 11.15 | 27.75 ± 6.47 | 31.74° N | 105.96° E |
| 蓬安县 PA | 30 | 109.95 ± 16.58 | 25.43 ± 33.82 | 31.04° N | 106.40° E |
| 合川区 HC | 28 | 114.84 ± 11.07 | 17.42 ± 4.40 | 29.98° N | 106.29° E |
| 位点 Loci | 引物序列 Primer sequence (5'-3') | 重复单元 Repeat motif | 片段大小 Size range (bp) | 退火温度 Tm (℃) | 参考来源 Reference source | 登录号 Accession number |
|---|---|---|---|---|---|---|
| SD01 | GCTCCATTCCCAACCTATAACACAT GCCTTGATGAACGGTTATGTATG | (ACAT)10 | 98 | 61 | sdF5010 | KX250317 |
| SD02 | GTGGCTCCTTCCCTTTCACAGAGCA GATTGTACATGTGGCTCAAG | (GCA)9 | 116 | 62 | sdF6400 | KX25347 |
| SD03 | ATGACAAAGGAAAACCACGAGAAA TGCAGATACAGCGCATCACTTTAG | (AAAC)16 | 132 | 62 | sdF5261 | KX250326 |
| SD04 | CACAGACGAGGATTCCAGTCGACA TGCAGTCCCTTACCTGTTCTC | (GAG)5 | 154 | 65 | sdF6273 | KX250343 |
| SD05 | TCCGTTGTTTAGGCTACTGATCAAA TGAGATGACATGACGATAGCTGTG | (GATA)17 | 155 | 63 | sdF6479 | KX250350 |
| SD07 | ACACTACTCGTCTGCCGCAAACAA TGCAGATTGTTTCAAAGCAG | (AAAT)2 | 111 | 53 | sdF7209 | KX250353 |
| SD08 | AACTGTAGGGCACGACAAATTGAT AGTCTAAACCCGTCTGCAAGAATG | (AGAC)18 | 232 | 56 | sdF5145 | KX250320 |
| SD09 | TCTCAGATGACGTTGAGCATATTGA CATTCATCTGGGCTCACTAAAACA | (ATCT)15 | 151 | 57 | sdF5163 | KX250321 |
| SD10 | TTTCTGTACTTGTTAGTTTGGGGTC AGATCAATTAAATGCATCATTGCTGA | (TTTC)6 | 156 | 56 | sdF5351 | KX250333 |
| SD11 | CGTCTAGTGCTGAAGGAGGTGAGT TCTCAGCCTGGAACACAGAGAGAT | (ACTC)25 | 123 | 62 | sdF6476 | KX25329 |
Table 2 Primer sequences and characteristics of microsatellite loci of Saurogobio dabryi
| 位点 Loci | 引物序列 Primer sequence (5'-3') | 重复单元 Repeat motif | 片段大小 Size range (bp) | 退火温度 Tm (℃) | 参考来源 Reference source | 登录号 Accession number |
|---|---|---|---|---|---|---|
| SD01 | GCTCCATTCCCAACCTATAACACAT GCCTTGATGAACGGTTATGTATG | (ACAT)10 | 98 | 61 | sdF5010 | KX250317 |
| SD02 | GTGGCTCCTTCCCTTTCACAGAGCA GATTGTACATGTGGCTCAAG | (GCA)9 | 116 | 62 | sdF6400 | KX25347 |
| SD03 | ATGACAAAGGAAAACCACGAGAAA TGCAGATACAGCGCATCACTTTAG | (AAAC)16 | 132 | 62 | sdF5261 | KX250326 |
| SD04 | CACAGACGAGGATTCCAGTCGACA TGCAGTCCCTTACCTGTTCTC | (GAG)5 | 154 | 65 | sdF6273 | KX250343 |
| SD05 | TCCGTTGTTTAGGCTACTGATCAAA TGAGATGACATGACGATAGCTGTG | (GATA)17 | 155 | 63 | sdF6479 | KX250350 |
| SD07 | ACACTACTCGTCTGCCGCAAACAA TGCAGATTGTTTCAAAGCAG | (AAAT)2 | 111 | 53 | sdF7209 | KX250353 |
| SD08 | AACTGTAGGGCACGACAAATTGAT AGTCTAAACCCGTCTGCAAGAATG | (AGAC)18 | 232 | 56 | sdF5145 | KX250320 |
| SD09 | TCTCAGATGACGTTGAGCATATTGA CATTCATCTGGGCTCACTAAAACA | (ATCT)15 | 151 | 57 | sdF5163 | KX250321 |
| SD10 | TTTCTGTACTTGTTAGTTTGGGGTC AGATCAATTAAATGCATCATTGCTGA | (TTTC)6 | 156 | 56 | sdF5351 | KX250333 |
| SD11 | CGTCTAGTGCTGAAGGAGGTGAGT TCTCAGCCTGGAACACAGAGAGAT | (ACTC)25 | 123 | 62 | sdF6476 | KX25329 |
| 种群 Populations | mtDNA线粒体控制区 mtDNA control region | 微卫星位点 Microsatellite loci | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | H | Hd | π | n | k | A | Ai | Ho | He | PIC | Fis | |
| 朝天区 CT | 27 | 18 | 0.929 | 0.0180 | 94 | 16.328 | 4.500 | 4.500 | 0.493 | 0.433 | 0.391 | -0.139 |
| 苍溪县 CX | 28 | 10 | 0.877 | 0.0100 | 43 | 9.050 | 4.600 | 5.711 | 0.273 | 0.378 | 0.302 | 0.152 |
| 蓬安县 PA | 30 | 14 | 0.875 | 0.0120 | 89 | 10.863 | 6.400 | 6.169 | 0.357 | 0.490 | 0.443 | 0.275 |
| 合川区 HC | 28 | 17 | 0.940 | 0.0264 | 129 | 23.818 | 8.700 | 8.319 | 0.483 | 0.579 | 0.543 | 0.150 |
| 总计 Total | 113 | 39 | 0.894 | 0.0348 | 203 | 12.654 | 12 | 5.923 | 0.302 | 0.438 | 0.531 | 0.145 |
Table 3 Genetic variability of Saurogobio dabryi populations based on mtDNA control region and microsatellite loci. CT, Chaotian; CX, Cangxi; PA, Peng’an; HC, Hechuan.
| 种群 Populations | mtDNA线粒体控制区 mtDNA control region | 微卫星位点 Microsatellite loci | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | H | Hd | π | n | k | A | Ai | Ho | He | PIC | Fis | |
| 朝天区 CT | 27 | 18 | 0.929 | 0.0180 | 94 | 16.328 | 4.500 | 4.500 | 0.493 | 0.433 | 0.391 | -0.139 |
| 苍溪县 CX | 28 | 10 | 0.877 | 0.0100 | 43 | 9.050 | 4.600 | 5.711 | 0.273 | 0.378 | 0.302 | 0.152 |
| 蓬安县 PA | 30 | 14 | 0.875 | 0.0120 | 89 | 10.863 | 6.400 | 6.169 | 0.357 | 0.490 | 0.443 | 0.275 |
| 合川区 HC | 28 | 17 | 0.940 | 0.0264 | 129 | 23.818 | 8.700 | 8.319 | 0.483 | 0.579 | 0.543 | 0.150 |
| 总计 Total | 113 | 39 | 0.894 | 0.0348 | 203 | 12.654 | 12 | 5.923 | 0.302 | 0.438 | 0.531 | 0.145 |
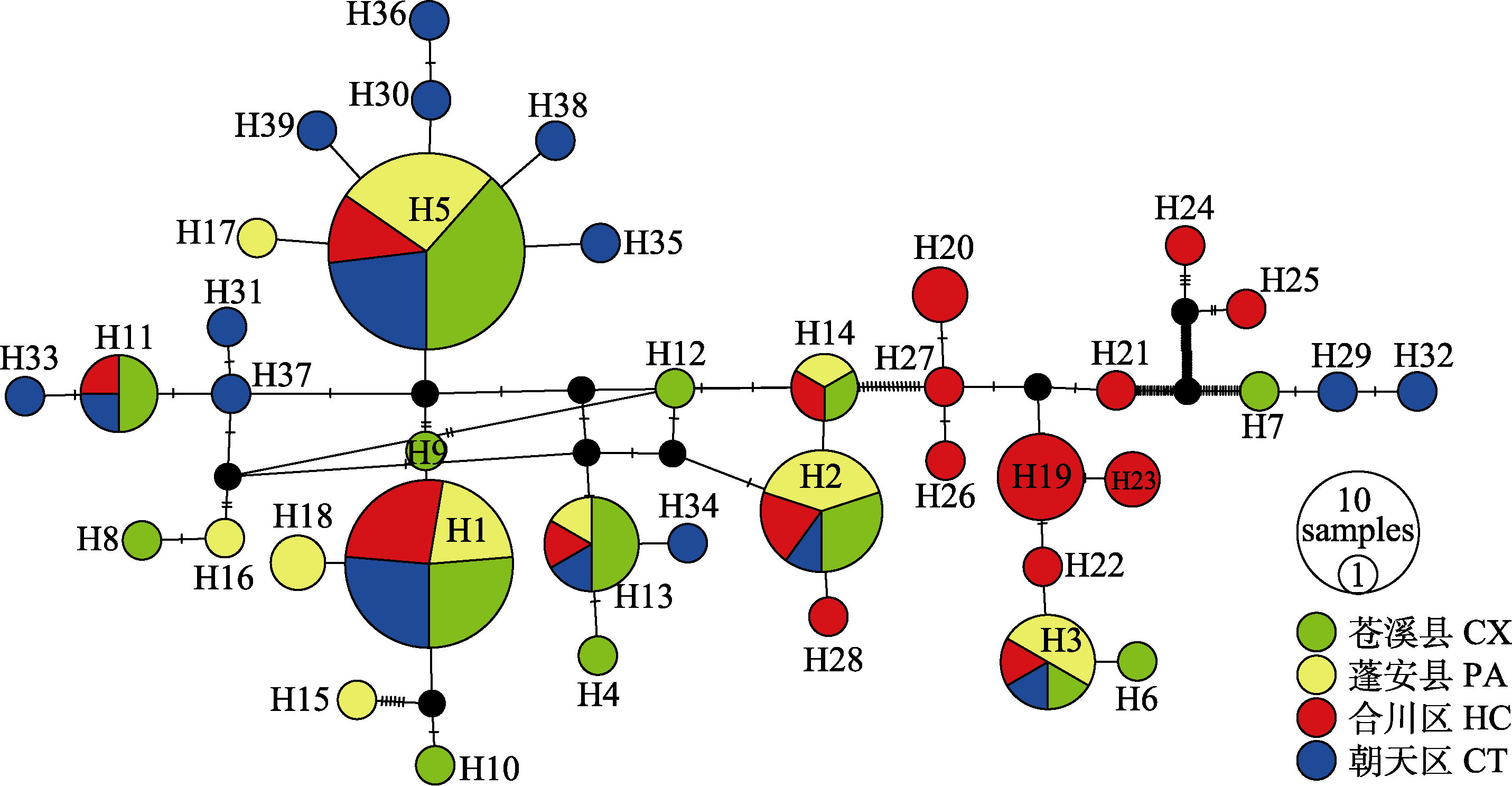
Fig. 2 Haplotype network for 39 haplotypes of four Saurogobio dabryi populations in Jialing River based on the median-joining algorithm. CT, Chaotian; CX, Cangxi; PA, Peng’an; HC, Hechuan.
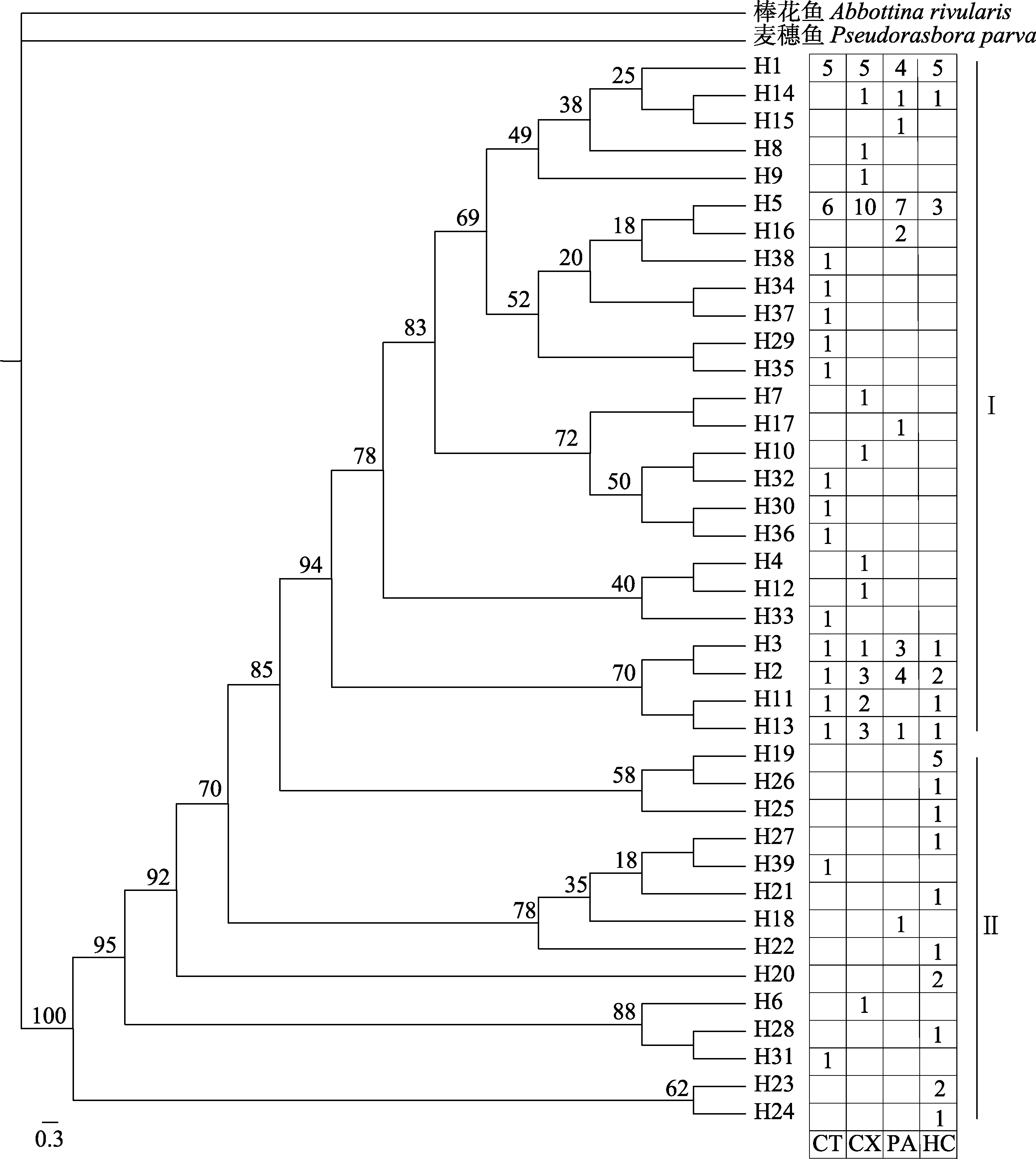
Fig. 3 Molecular phylogenetic tree of Saurogobio dabryi derived from the mitochondrial control region. The number of each branch correspond to the Bayesian posterior probability. CT, Chaotian; CX, Cangxi; PA, Peng’an; HC, Hechuan.
| 变异来源 Source of variation | 自由度 Degree of freedom | 总方差 Total variance | 变异比率 Variation ratio (%) | 固定系数 Fixed parameter | 概率 Probability |
|---|---|---|---|---|---|
| 组间 Intergroup | 2 | 733.641 | 69.38 | 0.02561 | 0.00027 ± 0.01111 |
| 组内种群间 Intragroup inter-population | 1 | 81.944 | 12.85 | 0.10621 | 0.00047 ± 0.01031 |
| 种群内个体间 Among individuals within a population | 110 | 102.266 | 22.23 | 0.12853 | 0.00000 ± 0.00000 |
| 合计 Total | 113 | 917.851 |
Table 4 The AMOVA analysis of the four Saurogobio dabryi populations
| 变异来源 Source of variation | 自由度 Degree of freedom | 总方差 Total variance | 变异比率 Variation ratio (%) | 固定系数 Fixed parameter | 概率 Probability |
|---|---|---|---|---|---|
| 组间 Intergroup | 2 | 733.641 | 69.38 | 0.02561 | 0.00027 ± 0.01111 |
| 组内种群间 Intragroup inter-population | 1 | 81.944 | 12.85 | 0.10621 | 0.00047 ± 0.01031 |
| 种群内个体间 Among individuals within a population | 110 | 102.266 | 22.23 | 0.12853 | 0.00000 ± 0.00000 |
| 合计 Total | 113 | 917.851 |
| 种群 Population | CT | CX | PA | HC |
|---|---|---|---|---|
| 朝天区 CT | 0.18921* | 0.01396** | 0.21069* | |
| 苍溪县 CX | 0.00028 | 0.03035* | 0.16243** | |
| 蓬安县 PA | 0.01624 | 0.01993 | 0.14526** | |
| 合川区 HC | 0.12140** | 0.13835* | 0.15537* |
Table 5 Pairwise genetic differentiation (Fst) of Saurogobio dabryi populations based on microsatellite loci (above the diagonal) and mtDNA control region (below the diagonal). CT, Chaotian; CX, Cangxi; PA, Peng’an; HC, Hechuan.
| 种群 Population | CT | CX | PA | HC |
|---|---|---|---|---|
| 朝天区 CT | 0.18921* | 0.01396** | 0.21069* | |
| 苍溪县 CX | 0.00028 | 0.03035* | 0.16243** | |
| 蓬安县 PA | 0.01624 | 0.01993 | 0.14526** | |
| 合川区 HC | 0.12140** | 0.13835* | 0.15537* |
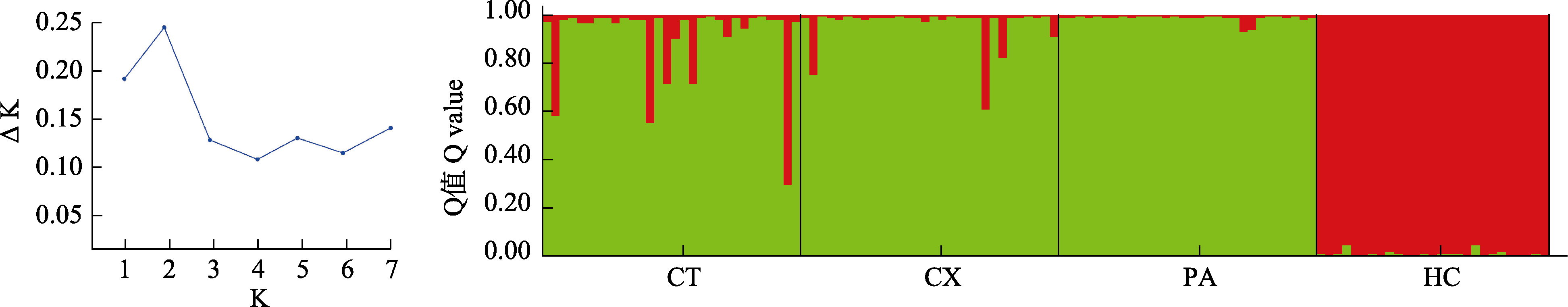
Fig. 4 Genetic structure of four Saurogobio dabryi populations based on K = 2. CT, Chaotian; CX, Cangxi; PA, Peng’an; HC, Hechuan. K, Number of subgroups. ΔK = mean(|L"(K)|) / sd(L(K)).
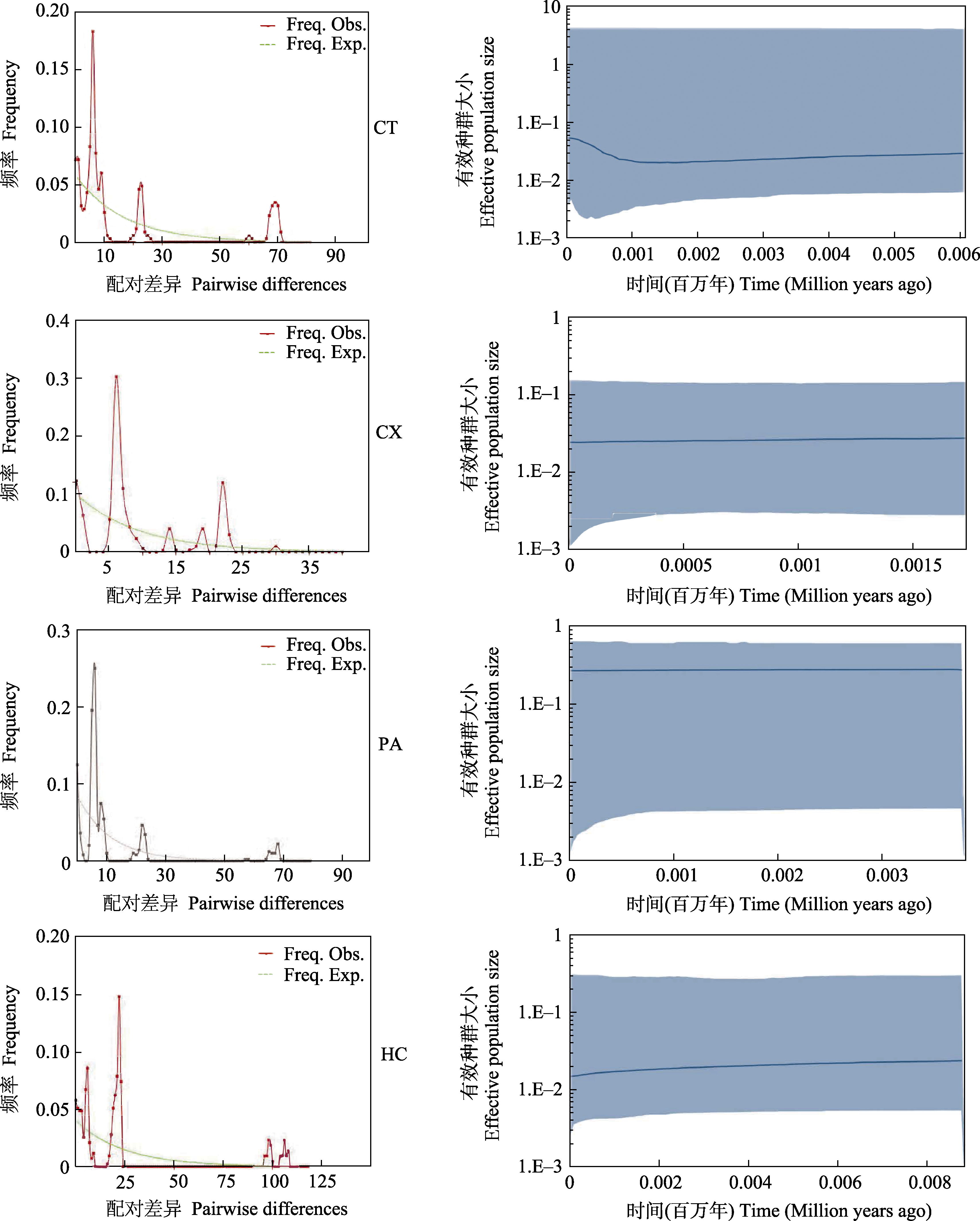
Fig. 5 Mismatch distributions and Bayesian skyline plot of mitochondrial lineages of four Saurogobio dabryi populations. Freq. Obs., Observed value; Freq. Exp., Expected value.
| 种群 Population | Afd | P(The/d) | P(OHe) | P(OHd) | 种群 Population | Afd | P(The/d) | P(OHe) | P(OHd) |
|---|---|---|---|---|---|---|---|---|---|
| 朝天区 CT | L-shaped | 0.64063 | 0.72656 | 0.01031 | 蓬安县 PA | L-shaped | 0.03711 | 0.98633 | 0.19855 |
| 苍溪县 CX | L-shaped | 0.01367 | 0.99512 | 0.30684 | 合川区 HC | L-shaped | 0.00977 | 0.99658 | 0.20488 |
Table 6 Bottleneck tests for microsatellite loci applied in Saurogobio dabryi populations
| 种群 Population | Afd | P(The/d) | P(OHe) | P(OHd) | 种群 Population | Afd | P(The/d) | P(OHe) | P(OHd) |
|---|---|---|---|---|---|---|---|---|---|
| 朝天区 CT | L-shaped | 0.64063 | 0.72656 | 0.01031 | 蓬安县 PA | L-shaped | 0.03711 | 0.98633 | 0.19855 |
| 苍溪县 CX | L-shaped | 0.01367 | 0.99512 | 0.30684 | 合川区 HC | L-shaped | 0.00977 | 0.99658 | 0.20488 |
| [1] |
Blanchet S, Rey O, Etienne R, Lek S, Loot G (2010) Species-specific responses to landscape fragmentation: Implications for management strategies. Evolutionary Applications, 3, 291-304.
DOI PMID |
| [2] | Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 10, e1003537. |
| [3] |
Bu QT, Li X, Zhu R, Chu L, Yan YZ (2017) Low-head dams driving the homogenization of local habitat and fish assemblages in upland streams of the Qingyi River. Biodiversity Science, 25, 830-839. (in Chinese with English abstract)
DOI |
|
[卜倩婷, 李献, 朱仁, 储玲, 严云志 (2017) 低头坝驱动山区溪流局域栖息地和鱼类群落的同质化. 生物多样性, 25, 830-839.]
DOI |
|
| [4] | Chen K, Luo MY, Zhang CK, Zhang DC, Xu WC (2007) Countermeasures to multistage navigable electricity project’s impaction on the Jialing River (Nanchong) to ecological environment. Journal of China West Normal University (Natural Sciences), 28, 195-199. (in Chinese with English abstract) |
| [谌柯, 罗明云, 张崇昆, 张道川, 许武成 (2007) 嘉陵江干流(南充段)梯级航电开发对生态环境的影响和对策研究. 西华师范大学学报(自然科学版), 28, 195-199.] | |
| [5] | Chen YY (1998) Fauna Sinica·Osteichthyes·Cypriniformes (Ⅱ). Science Press, Beijing.. (in Chinese) |
| [陈宜瑜 (1998) 中国动物志·硬骨鱼纲·鲤形目(中卷). 科学出版社, 北京.] | |
| [6] |
Cole TL, Hammer MP, Unmack PJ, Teske PR, Brauer CJ, Adams M, Beheregaray LB (2016) Range-wide fragmentation in a threatened fish associated with post-European settlement modification in the Murray-Darling Basin, Australia. Conservation Genetics, 17, 1377-1391.
DOI |
| [7] |
de Brito MTFM, Leila SAT, da Silva Pinto D (2018) Seroepidemiology of arbovirus in communities living under the influence of the lake of a hydroelectric dam in Brazil. Cadernos Saúde Coletiva, 26, 1-6.
DOI URL |
| [8] | Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics, 1, 47-50. |
| [9] |
Faulks LK, Kerezsy A, Unmack PJ, Johnson JB, Hughes JM (2017) Going, going, gone? Loss of genetic diversity in two critically endangered Australian freshwater fishes, Scaturiginichthys vermeilipinnis and Chlamydogobius squamigenus, from Great Artesian Basin springs at Edgbaston, Queensland, Australia. Aquatic Conservation: Marine and Freshwater Ecosystems, 27, 39-50.
DOI URL |
| [10] | Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW (1995) Genetic absolute dating based on microsatellites and the origin of modern humans. Proceedings of the National Academy of Sciences, USA, 92, 6723-6727. |
| [11] |
Gouskov A, Reyes M, Wirthner-Bitterlin L, Vorburger C (2016) Fish population genetic structure shaped by hydroelectric power plants in the upper Rhine Catchment. Evolutionary Applications, 9, 394-408.
DOI PMID |
| [12] | He P, Jiang ZB, Yu SD, Hu J (2003) Water pollution evaluation under present condition and its control measures in Jialing River of Nanchong Section. Sichuan Journal of Environment, 22(3), 35-37. (in Chinese with English abstract) |
| [何平, 蒋祖斌, 余世东, 胡健 (2003) 嘉陵江南充段水污染现状评价及防治对策. 四川环境, 22(3), 35-37.] | |
| [13] | He XF, Song ZB, Xie EY (1996) The breeding habits and embryonic development of longnose gudgeon (Saurogobio dabryi Bleeker). Journal of China West Normal University (Natural Sciences), 21, 276-281. (in Chinese with English abstract) |
| [何学福, 宋昭彬, 谢恩义 (1996) 蛇鮈的产卵习性及胚胎发育. 西南师范大学学报(自然科学版), 21, 276-281.] | |
| [14] |
Heilveil JS, Stockwell CA (2017) Genetic signatures of translocations and habitat fragmentation for two evolutionarily significant units of a protected fish species. Environmental Biology of Fishes, 100, 631-638.
DOI URL |
| [15] | Hou FX (2013) The Genetic Diversity and Population Structure Analysis of Schizopygopsis chengi and Schizopygopsis malacanthus. PhD dissertation, Sichuan University, Chengdu. (in Chinese with English abstract) |
| [侯飞侠 (2013) 大渡裸裂尻鱼和软刺裸裂尻鱼的遗传多样性及种群结构研究. 博士学位论文, 四川大学, 成都.] | |
| [16] | Jiang GF, He XF (2008) Status of fish resources in the lower reaches of the Jialing River. Freshwater Fisheries, 38(2), 3-7. (in Chinese with English abstract) |
| [蒋国福, 何学福 (2008) 嘉陵江下游鱼类资源现状调查. 淡水渔业, 38(2), 3-7.] | |
| [17] |
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16, 1099-1106.
DOI PMID |
| [18] |
Khaire D, Atkulwar A, Farah S, Baig M (2017) Mitochondrial DNA analyses revealed low genetic diversity in the endangered Indian wild ass Equus hemionus khur. Mitochondrial DNA: Part A, 28, 681-686.
DOI URL |
| [19] |
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874.
DOI PMID |
| [20] |
Leigh JW, Bryant D (2015) Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110-1116.
DOI URL |
| [21] | Liu HG, Yang Z, Tang HY, Gong Y, Wan L (2017) Microsatellite development and characterization for Saurogobio dabryi Bleeker, 1871 in a Yangtze River- connected Lake, China. Journal of Genetics, 96, e1-e4. |
| [22] |
Liu HZ, Tzeng CS, Teng HY (2002) Sequence variations in the mitochondrial DNA control region and their implications for the phylogeny of the Cypriniformes. Canadian Journal of Zoology, 80, 569-581.
DOI URL |
| [23] | Liu XL, Sun J, Han JQ, Wang YN, Tan JD (2019) Genetic variation of Rattus losea populations in island habitats. Acta Ecologica Sinica, 39, 6898-6907. (in Chinese with English abstract) |
| [刘小丽, 孙佼, 韩金巧, 王艳妮, 谭江东 (2019) 岛屿生境下黄毛鼠种群的遗传变异. 生态学报, 39, 6898-6907.] | |
| [24] | Liu YY, Zeng Y, Xiong XQ, Lü ZY, Hu Y, Jiang ZM, Cheng L, Yu DL (2019) Difference and adaptation of reproductive biological characteristics of Saurogobio dabryi in Jialing River cascade water conservancy project. Freshwater Fisheries, 49(2), 3-8. (in Chinese with English abstract) |
| [刘玉莹, 曾燏, 熊小琴, 吕振宇, 胡月, 蒋朝明, 程磊, 俞丹莉 (2019) 嘉陵江梯级水利工程开发下不同江段蛇鮈的繁殖生物学特性差异及适应. 淡水渔业, 49(2), 3-8.] | |
| [25] |
Pavlova A, Beheregaray LB, Coleman R, Gilligan D, Harrisson KA, Ingram BA, Kearns J, Lamb AM, Lintermans M, Lyon J, Nguyen TTT, Sasaki M, Tonkin Z, Yen JDL, Sunnucks P (2017) Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: A call for assisted gene flow. Evolutionary Applications, 10, 531-550.
DOI PMID |
| [26] | Posada D (2009) Selection of models of DNA evolution with jModelTest. Bioinformatics for DNA Sequence Analysis, 93-112. |
| [27] | Ren LP (2012) The Study of Muli-scale Health Evaluation of the Cascade Hydropower Development on the Jialing River in Sichuan. PhD dissertation, Chongqing University, Chongqing. (in Chinese with English abstract) |
| [任丽平 (2012) 嘉陵江(四川段)梯级开发的多尺度健康评价研究. 博士学位论文, 重庆大学, 重庆.] | |
| [28] |
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569.
DOI PMID |
| [29] |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574.
DOI PMID |
| [30] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299-3302.
DOI PMID |
| [31] |
Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics, 152, 1079-1089.
DOI PMID |
| [32] |
Sungirai M, Baron S, van der Merwe NA, Moyo DZ, De Clercq P, Maritz-Olivier C, Madder M (2018) Population structure and genetic diversity of Rhipicephalus microplus in Zimbabwe. Acta Tropica, 180, 42-46.
DOI URL |
| [33] | Tao JP, Gong YT, Tan XC, Yang Z, Chang JB (2012) Spatiotemporal patterns of the fish assemblages downstream of the Gezhouba Dam on the Yangtze River. Scientia Sinica (Vitae), 42, 677-688. (in Chinese with English abstract) |
| [陶江平, 龚昱田, 谭细畅, 杨志, 常剑波 (2012) 长江葛洲坝坝下江段鱼类群落变化的时空特征. 中国科学: 生命科学, 42, 677-688.] | |
| [34] | Tian HW, Duan XB, Wang DQ, Liu SP, Chen DQ (2013) Sequence variability of cytochrome b and genetic structure of Leptobotia elongata in the Upper Yangtze River. Freshwater Fisheries, 43, 13-18, 28. (in Chinese with English abstract) |
| [田辉伍, 段辛斌, 汪登强, 刘绍平, 陈大庆 (2013) 长江上游长薄鳅Cytb基因的序列变异与遗传结构分析. 淡水渔业, 43, 13-18, 28.] | |
| [35] | Turan C, Gurlek M, Erguden D, Yaglioglu D, Uyan A, Reyhaniye AN, Ozbalcilar B, Ozturk B, Erdogan ZA, Ivanova P, Soldo A (2015) Population genetic analysis of Atlantic bonito Sarda sarda (Bloch, 1793) using sequence analysis of mtDNA D-loop region. Fresenius Environmental Bulletin, 45, 231-237. |
| [36] |
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535-538.
DOI URL |
| [37] | Wang T, Du YY, Yang ZY, Zhang YP, Lou ZY, Jiao WL (2017) Population genetic structure of Schizopygopsis kialingensis inferred from mitochondrial D-loop sequences. Acta Ecologica Sinica, 37, 7741-7749. (in Chinese with English abstract) |
| [王太, 杜岩岩, 杨濯羽, 张艳萍, 娄忠玉, 焦文龙 (2017) 基于线粒体控制区的嘉陵裸裂尻鱼种群遗传结构分析. 生态学报, 37, 7741-7749.] | |
| [38] | Yang JQ, Hu XL, Tang WQ, Lin HD (2008) mtDNA control region sequence variation and genetic diversity of Coilia nasus in Yangtze River Estuary and its adjacent waters. Chinese Journal of Zoology, 43, 8-15. (in Chinese with English abstract) |
| [杨金权, 胡雪莲, 唐文乔, 林弘都 (2008) 长江口邻近水域刀鲚的线粒体控制区序列变异与遗传多样性. 动物学杂志, 43, 8-15.] | |
| [39] | Zeng Y, Chen YB, Li ZJ (2014) Utilization and protection status of fish resources in Jialing River. Tianjin Agricultural Sciences, 20, 60-62. (in Chinese with English abstract) |
| [曾燏, 陈永柏, 李钟杰 (2014) 嘉陵江鱼类资源利用与保护现状. 天津农业科学, 20, 60-62.] | |
| [40] | Zhang F, Zhang BW, Tang WQ, Liu J, Wu JH, Tang B (2018) Analysis of genetic diversity and population dynamics of the narrow-ridged finless porpoise in the Yangtze River Estuary. Journal of Shanghai Ocean University, 27, 656-665. (in Chinese with English abstract) |
| [张枫, 张保卫, 唐文乔, 刘健, 吴建辉, 唐斌 (2018) 长江口江豚的遗传多样性现状及种群动态. 上海海洋大学学报, 27, 656-665.] | |
| [41] | Zhang FT, Tan DQ (2010) Genetic diversity in population of largemouth bronze gudgeon (Coreius guichenoti Sauvage et Dabry) from Yangtze River determined by microsatellite DNA analysis. Genes & Genetic Systems, 85, 351-357. |
| [42] |
Zhou Y, Lei Y, Lu Y, Song ZB (2018) Population genetics of a Chinese endemic, Gymnocypris potanini Herzenstein, threatened by population isolation: Conflicting patterns between microsatellites and mitochondrial DNA. Hydrobiologia, 819, 145-159.
DOI |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [11] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [12] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [13] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| [14] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| [15] | Jun Sun, Yuyao Song, Yifeng Shi, Jian Zhai, Wenzhuo Yan. Progress of marine biodiversity studies in China seas in the past decade [J]. Biodiv Sci, 2022, 30(10): 22526-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()