



Biodiv Sci ›› 2021, Vol. 29 ›› Issue (12): 1629-1637. DOI: 10.17520/biods.2021341 cstr: 32101.14.biods.2021341
• Original Papers: Plant Diversity • Previous Articles Next Articles
Received:2021-08-30
Accepted:2021-11-19
Online:2021-12-20
Published:2021-12-16
Contact:
Bin Tian
Junwei Ye, Bin Tian. Genetic structure and its causes of an important woody oil plant in Southwest China, Prinsepia utilis (Rosaceae)[J]. Biodiv Sci, 2021, 29(12): 1629-1637.
| 代号 Code | 地点 Site | 经纬度 Location | 海拔 Altitude (m) | n | AO | HE | HO | RS | PAR | FIS |
|---|---|---|---|---|---|---|---|---|---|---|
| BAZY | 云南昆明 Kunming, Yunnan | 102.72˚ E, 25.40˚ N | 2,370 | 9 | 20 | 0.45 | 0.63 | 2.45 | 0.01 | -0.35 |
| BBM | 西藏波密 Bomi, Xizang | 95.34˚ E, 29.99˚ N | 2,750 | 14 | 22 | 0.54 | 0.76 | 2.61 | 0 | -0.37 |
| BDC | 四川稻城 Daocheng, Sichuan | 100.30˚ E, 28.58˚ N | 3,310 | 12 | 21 | 0.46 | 0.62 | 2.55 | 0.12 | -0.31 |
| BDCQ | 云南会泽 Huize, Yunnan | 103.10˚ E, 25.94˚ N | 2,220 | 10 | 22 | 0.48 | 0.67 | 2.74 | 0.07 | -0.35 |
| BES | 云南峨山 E’shan, Yunnan | 102.11˚ E, 24.28˚ N | 2,060 | 6 | 15 | 0.38 | 0.62 | - | - | -0.58 |
| BHD | 四川会东 Huidong, Sichuan | 102.78˚ E, 26.55˚ N | 2,370 | 14 | 22 | 0.47 | 0.65 | 2.61 | 0 | -0.36 |
| BHQ | 云南鹤庆 Heqing, Yunnan | 100.18˚ E, 26.56˚ N | 2,200 | 15 | 27 | 0.64 | 0.86 | 3.28 | 0 | -0.31 |
| BJL | 西藏吉隆 Jilong, Xizang | 85.36˚ E, 28.39˚ N | 3,140 | 12 | 19 | 0.46 | 0.65 | 2.40 | 0.03 | -0.38 |
| BJP | 云南澄江 Chengjiang, Yunnan | 102.86˚ E, 24.73˚ N | 2,180 | 15 | 25 | 0.58 | 0.81 | 2.93 | 0 | -0.38 |
| BLD | 云南玉龙 Yulong, Yunnan | 99.46˚ E, 27.18˚ N | 2,630 | 16 | 21 | 0.56 | 0.88 | 2.59 | 0 | -0.54 |
| BLJ | 云南丽江 Lijiang, Yunnan | 100.23˚ E, 27.18˚ N | 3,650 | 5 | 19 | 0.52 | 0.78 | - | - | -0.47 |
| BLJS | 云南老君山 Laojun Mountains, Yunnan | 103.93˚ E, 23.30˚ N | 1,640 | 15 | 25 | 0.55 | 0.84 | 2.83 | 0 | -0.51 |
| BLJW | 贵州安顺 Anshun, Guizhou | 105.94˚ E, 26.25˚ N | 1,400 | 15 | 23 | 0.49 | 0.69 | 2.63 | 0.10 | -0.36 |
| BLLX | 贵州龙里 Longli, Guizhou | 106.90˚ E, 26.43˚ N | 1,150 | 10 | 18 | 0.45 | 0.61 | 2.30 | 0 | -0.37 |
| BLP | 云南兰坪 Lanping, Yunnan | 99.42˚ E, 26.46˚ N | 2,540 | 12 | 24 | 0.61 | 0.98 | 2.93 | 0 | -0.57 |
| BMG | 云南马关 Maguan, Yunnan | 104.39˚ E, 23.02˚ N | 1,340 | 12 | 24 | 0.53 | 0.79 | 2.80 | 0.02 | -0.45 |
| BML | 四川木里 Muli, Sichuan | 101.25˚ E, 27.97˚ N | 2,500 | 12 | 26 | 0.61 | 0.96 | 3.05 | 0.19 | -0.55 |
| BNH | 云南南华 Nanhua, Yunnan | 101.01˚ E, 25.33˚ N | 2,320 | 9 | 21 | 0.52 | 0.75 | 2.73 | 0 | -0.39 |
| BQJ | 云南曲靖 Qujing, Yunnan | 103.91˚ E, 25.36˚ N | 1,900 | 8 | 20 | 0.49 | 0.74 | - | - | -0.44 |
| BQL | 贵州贵阳 Guiyang, Guizhou | 106.69˚ E, 26.60˚ N | 1,160 | 6 | 17 | 0.45 | 0.60 | - | - | -0.24 |
| BSZ | 云南师宗 Shizong, Yunnan | 103.98˚ E, 24.82˚ N | 1,870 | 12 | 25 | 0.52 | 0.71 | 2.91 | 0.03 | -0.34 |
| BTM | 西藏林芝 Linzhi, Xizang | 96.58˚ E, 30.50˚ N | 4,720 | 15 | 25 | 0.59 | 0.85 | 2.83 | 0.09 | -0.41 |
| BWM | 云南禄劝 Luquan, Yunnan | 102.83˚ E, 26.02˚ N | 2,760 | 14 | 22 | 0.47 | 0.69 | 2.49 | 0 | -0.46 |
| BWN | 贵州威宁 Weining, Guizhou | 104.11˚ E, 26.74˚ N | 2,450 | 13 | 24 | 0.48 | 0.68 | 2.61 | 0 | -0.38 |
| BWX | 云南维西 Weixi, Yunnan | 99.02˚ E, 27.80˚ N | 2,290 | 9 | 24 | 0.59 | 0.87 | 2.97 | 0.05 | -0.43 |
| BXC | 四川西昌 Xichang, Sichuan | 102.35˚ E, 27.70˚ N | 2,680 | 11 | 20 | 0.39 | 0.57 | 2.38 | 0.03 | -0.41 |
| BXCX | 云南西畴 Xichou, Yunnan | 104.79˚ E, 23.38˚ N | 1,450 | 9 | 25 | 0.53 | 0.75 | 2.86 | 0 | -0.37 |
| BXJ | 云南文山新街 Xinjie, Wenshan, Yunnan | 104.00˚ E, 23.20˚ N | 1,880 | 15 | 23 | 0.53 | 0.81 | 2.76 | 0 | -0.49 |
| BXZC | 云南开远 Kaiyuan, Yunnan | 103.62˚ E, 23.79˚ N | 1,480 | 10 | 21 | 0.54 | 0.80 | 2.77 | 0 | -0.43 |
| BYG | 西藏波密 Bomi, Xizang | 95.57˚ E, 30.50˚ N | 5,230 | 15 | 22 | 0.58 | 0.88 | 2.79 | 0.01 | -0.48 |
| BYXB | 四川越西 Yuexi, Sichuan | 102.45˚ E, 28.76˚ N | 2,970 | 12 | 16 | 0.39 | 0.60 | 2.12 | 0 | -0.49 |
| BZNX | 云南文山 Wenshan, Yunnan | 104.42˚ E, 23.69˚ N | 1,570 | 15 | 25 | 0.55 | 0.81 | 2.97 | 0 | -0.45 |
Table 1 Sampling and genetic diversity information of Prinsepia utilis
| 代号 Code | 地点 Site | 经纬度 Location | 海拔 Altitude (m) | n | AO | HE | HO | RS | PAR | FIS |
|---|---|---|---|---|---|---|---|---|---|---|
| BAZY | 云南昆明 Kunming, Yunnan | 102.72˚ E, 25.40˚ N | 2,370 | 9 | 20 | 0.45 | 0.63 | 2.45 | 0.01 | -0.35 |
| BBM | 西藏波密 Bomi, Xizang | 95.34˚ E, 29.99˚ N | 2,750 | 14 | 22 | 0.54 | 0.76 | 2.61 | 0 | -0.37 |
| BDC | 四川稻城 Daocheng, Sichuan | 100.30˚ E, 28.58˚ N | 3,310 | 12 | 21 | 0.46 | 0.62 | 2.55 | 0.12 | -0.31 |
| BDCQ | 云南会泽 Huize, Yunnan | 103.10˚ E, 25.94˚ N | 2,220 | 10 | 22 | 0.48 | 0.67 | 2.74 | 0.07 | -0.35 |
| BES | 云南峨山 E’shan, Yunnan | 102.11˚ E, 24.28˚ N | 2,060 | 6 | 15 | 0.38 | 0.62 | - | - | -0.58 |
| BHD | 四川会东 Huidong, Sichuan | 102.78˚ E, 26.55˚ N | 2,370 | 14 | 22 | 0.47 | 0.65 | 2.61 | 0 | -0.36 |
| BHQ | 云南鹤庆 Heqing, Yunnan | 100.18˚ E, 26.56˚ N | 2,200 | 15 | 27 | 0.64 | 0.86 | 3.28 | 0 | -0.31 |
| BJL | 西藏吉隆 Jilong, Xizang | 85.36˚ E, 28.39˚ N | 3,140 | 12 | 19 | 0.46 | 0.65 | 2.40 | 0.03 | -0.38 |
| BJP | 云南澄江 Chengjiang, Yunnan | 102.86˚ E, 24.73˚ N | 2,180 | 15 | 25 | 0.58 | 0.81 | 2.93 | 0 | -0.38 |
| BLD | 云南玉龙 Yulong, Yunnan | 99.46˚ E, 27.18˚ N | 2,630 | 16 | 21 | 0.56 | 0.88 | 2.59 | 0 | -0.54 |
| BLJ | 云南丽江 Lijiang, Yunnan | 100.23˚ E, 27.18˚ N | 3,650 | 5 | 19 | 0.52 | 0.78 | - | - | -0.47 |
| BLJS | 云南老君山 Laojun Mountains, Yunnan | 103.93˚ E, 23.30˚ N | 1,640 | 15 | 25 | 0.55 | 0.84 | 2.83 | 0 | -0.51 |
| BLJW | 贵州安顺 Anshun, Guizhou | 105.94˚ E, 26.25˚ N | 1,400 | 15 | 23 | 0.49 | 0.69 | 2.63 | 0.10 | -0.36 |
| BLLX | 贵州龙里 Longli, Guizhou | 106.90˚ E, 26.43˚ N | 1,150 | 10 | 18 | 0.45 | 0.61 | 2.30 | 0 | -0.37 |
| BLP | 云南兰坪 Lanping, Yunnan | 99.42˚ E, 26.46˚ N | 2,540 | 12 | 24 | 0.61 | 0.98 | 2.93 | 0 | -0.57 |
| BMG | 云南马关 Maguan, Yunnan | 104.39˚ E, 23.02˚ N | 1,340 | 12 | 24 | 0.53 | 0.79 | 2.80 | 0.02 | -0.45 |
| BML | 四川木里 Muli, Sichuan | 101.25˚ E, 27.97˚ N | 2,500 | 12 | 26 | 0.61 | 0.96 | 3.05 | 0.19 | -0.55 |
| BNH | 云南南华 Nanhua, Yunnan | 101.01˚ E, 25.33˚ N | 2,320 | 9 | 21 | 0.52 | 0.75 | 2.73 | 0 | -0.39 |
| BQJ | 云南曲靖 Qujing, Yunnan | 103.91˚ E, 25.36˚ N | 1,900 | 8 | 20 | 0.49 | 0.74 | - | - | -0.44 |
| BQL | 贵州贵阳 Guiyang, Guizhou | 106.69˚ E, 26.60˚ N | 1,160 | 6 | 17 | 0.45 | 0.60 | - | - | -0.24 |
| BSZ | 云南师宗 Shizong, Yunnan | 103.98˚ E, 24.82˚ N | 1,870 | 12 | 25 | 0.52 | 0.71 | 2.91 | 0.03 | -0.34 |
| BTM | 西藏林芝 Linzhi, Xizang | 96.58˚ E, 30.50˚ N | 4,720 | 15 | 25 | 0.59 | 0.85 | 2.83 | 0.09 | -0.41 |
| BWM | 云南禄劝 Luquan, Yunnan | 102.83˚ E, 26.02˚ N | 2,760 | 14 | 22 | 0.47 | 0.69 | 2.49 | 0 | -0.46 |
| BWN | 贵州威宁 Weining, Guizhou | 104.11˚ E, 26.74˚ N | 2,450 | 13 | 24 | 0.48 | 0.68 | 2.61 | 0 | -0.38 |
| BWX | 云南维西 Weixi, Yunnan | 99.02˚ E, 27.80˚ N | 2,290 | 9 | 24 | 0.59 | 0.87 | 2.97 | 0.05 | -0.43 |
| BXC | 四川西昌 Xichang, Sichuan | 102.35˚ E, 27.70˚ N | 2,680 | 11 | 20 | 0.39 | 0.57 | 2.38 | 0.03 | -0.41 |
| BXCX | 云南西畴 Xichou, Yunnan | 104.79˚ E, 23.38˚ N | 1,450 | 9 | 25 | 0.53 | 0.75 | 2.86 | 0 | -0.37 |
| BXJ | 云南文山新街 Xinjie, Wenshan, Yunnan | 104.00˚ E, 23.20˚ N | 1,880 | 15 | 23 | 0.53 | 0.81 | 2.76 | 0 | -0.49 |
| BXZC | 云南开远 Kaiyuan, Yunnan | 103.62˚ E, 23.79˚ N | 1,480 | 10 | 21 | 0.54 | 0.80 | 2.77 | 0 | -0.43 |
| BYG | 西藏波密 Bomi, Xizang | 95.57˚ E, 30.50˚ N | 5,230 | 15 | 22 | 0.58 | 0.88 | 2.79 | 0.01 | -0.48 |
| BYXB | 四川越西 Yuexi, Sichuan | 102.45˚ E, 28.76˚ N | 2,970 | 12 | 16 | 0.39 | 0.60 | 2.12 | 0 | -0.49 |
| BZNX | 云南文山 Wenshan, Yunnan | 104.42˚ E, 23.69˚ N | 1,570 | 15 | 25 | 0.55 | 0.81 | 2.97 | 0 | -0.45 |
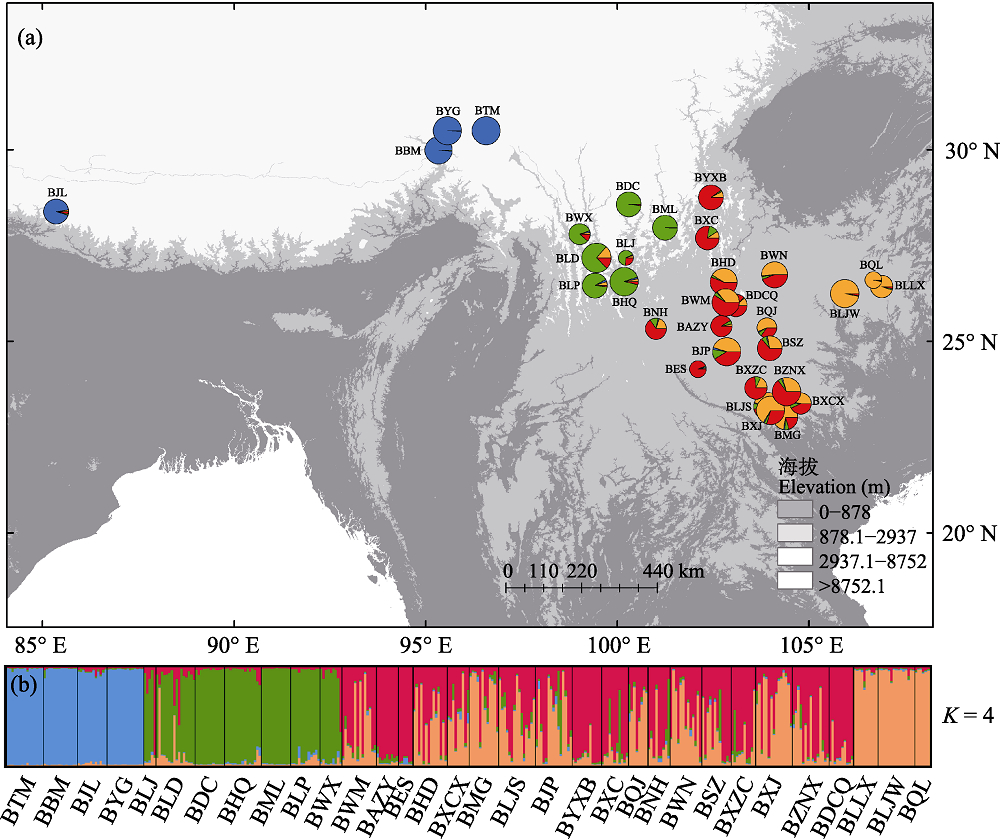
Fig. 1 Color-coded clustering (a) and histogram of the cluster assignment of different individuals (b) of the 32 Prinsepia utilis populations when K = 4. Blue represents Himalayas group, green represents Hengduan Mountains group, red and orange represent west and east Yunnan-Guizhou Plateau group, respectively. Population codes are the same as that in Table 1.
| 遗传群组 Genetic cluster | 等位基因数量 AO | 观测杂合度 HO | 期望杂合度 HE | 等位基因丰富度 RS | 私有等位基因丰富度 PAR |
|---|---|---|---|---|---|
| 喜马拉雅 Himalayas | 30 | 0.79 | 0.62 | 3.18 | 0.70 |
| 横断山 Hengduan Mountains | 35 | 0.86 | 0.65 | 3.39 | 0.53 |
| 云贵高原西部 West Yunnan-Guizhou Plateau | 36 | 0.73 | 0.56 | 3.04 | 0.19 |
| 云贵高原东部 East Yunnan-Guizhou Plateau | 26 | 0.65 | 0.50 | 2.69 | 0.24 |
Table 2 Genetic diversity of different genetic cluster of Prinsepia utilis
| 遗传群组 Genetic cluster | 等位基因数量 AO | 观测杂合度 HO | 期望杂合度 HE | 等位基因丰富度 RS | 私有等位基因丰富度 PAR |
|---|---|---|---|---|---|
| 喜马拉雅 Himalayas | 30 | 0.79 | 0.62 | 3.18 | 0.70 |
| 横断山 Hengduan Mountains | 35 | 0.86 | 0.65 | 3.39 | 0.53 |
| 云贵高原西部 West Yunnan-Guizhou Plateau | 36 | 0.73 | 0.56 | 3.04 | 0.19 |
| 云贵高原东部 East Yunnan-Guizhou Plateau | 26 | 0.65 | 0.50 | 2.69 | 0.24 |
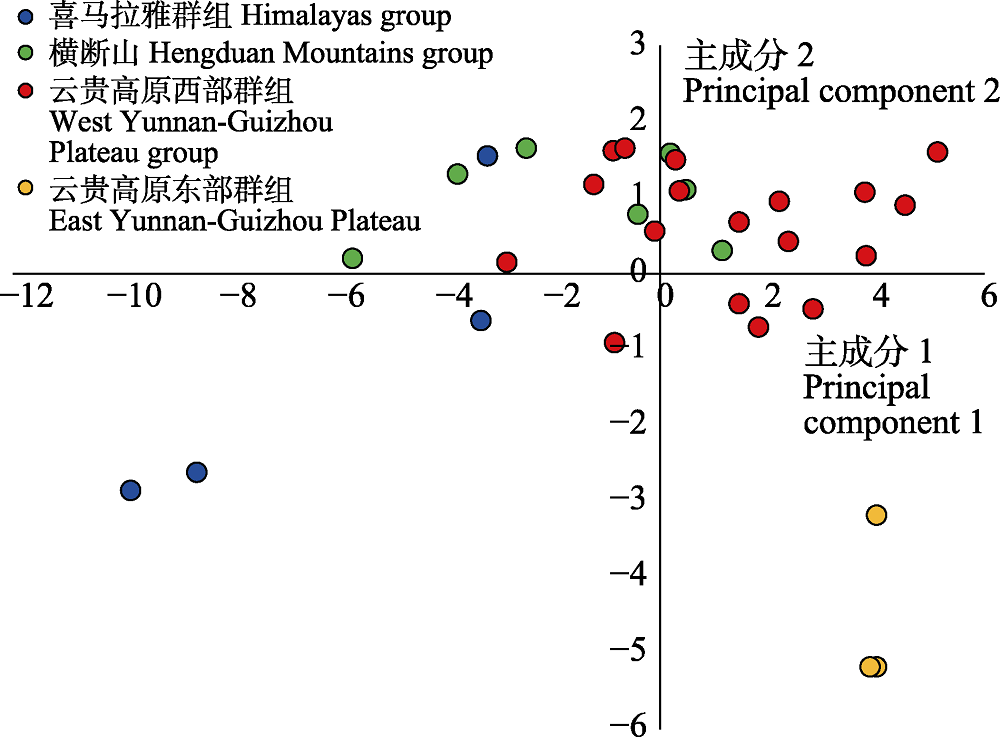
Fig. 2 Principal component analysis of climate variables of 32 Prinsepia utilis populations. Coded colors of different genetic clusters correspond to those in Fig. 1.
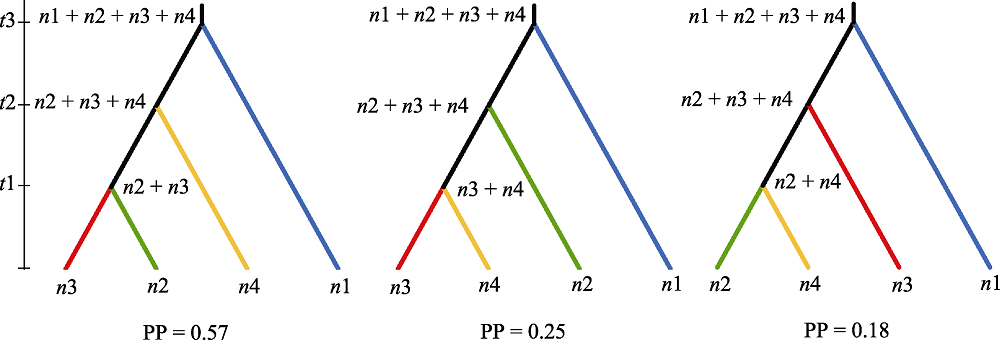
Fig. 3 Illustration of three different scenarios of divergence history of Prinsepia utilis populations. n1-4 represent effective population sizes of Himalayas, Hengduan Mountains, west and east Yunnan-Guizhou Plateau genetic groups, respectively; t1-3 represent divergence time of different divergence events (in generations); and PP represents posterior probability.
| n1 | n2 | n3 | n4 | T1 | T2 | T3 | μ | P | |
|---|---|---|---|---|---|---|---|---|---|
| 中位值 Median | 6.84×104 | 6.30×104 | 5.84×104 | 3.90×104 | 8.53×104 | 1.25×105 | 5.11×105 | 4.30×10-6 | 0.21 |
| 95%置信区间下限 95% HPD lower | 3.30×104 | 2.68×104 | 2.39×104 | 1.21×104 | 1.80×104 | 2.73×104 | 1.92×105 | 2.19×10-6 | 0.11 |
| 95%置信区间上限 95% HPD upper | 9.53×104 | 9.44×104 | 9.15×104 | 8.50×104 | 3.49×105 | 4.49×105 | 8.82×105 | 1.01×10-5 | 0.70 |
Table 3 Posterior median and 95% highest posterior density interval (HPD) estimate for demographic parameters in the most possible divergence scenario of Prinsepia utilis in DIYABC
| n1 | n2 | n3 | n4 | T1 | T2 | T3 | μ | P | |
|---|---|---|---|---|---|---|---|---|---|
| 中位值 Median | 6.84×104 | 6.30×104 | 5.84×104 | 3.90×104 | 8.53×104 | 1.25×105 | 5.11×105 | 4.30×10-6 | 0.21 |
| 95%置信区间下限 95% HPD lower | 3.30×104 | 2.68×104 | 2.39×104 | 1.21×104 | 1.80×104 | 2.73×104 | 1.92×105 | 2.19×10-6 | 0.11 |
| 95%置信区间上限 95% HPD upper | 9.53×104 | 9.44×104 | 9.15×104 | 8.50×104 | 3.49×105 | 4.49×105 | 8.82×105 | 1.01×10-5 | 0.70 |
| [1] | Bai WN, Zhang DY (2014) Current status and future directions in plant phylogeography. Chinese Bulletin of Life Sciences, 26, 125-137. (in Chinese with English abstract) |
| [ 白伟宁, 张大勇 (2014) 植物亲缘地理学的研究现状与发展趋势. 生命科学, 26, 125-137.] | |
| [2] |
Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics, 30, 1187-1189.
DOI URL |
| [3] |
Ding WN, Ree RH, Spicer RA, Xing YW (2020) Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science, 369, 578-581.
DOI URL |
| [4] |
Doyle JJ, Doyle JL (1990) Isolation of Plant DNA from fresh tissue. Focus, 12, 13-15.
DOI URL |
| [5] |
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611-2620.
PMID |
| [6] |
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Molecular Ecology Notes, 7, 574-578.
DOI URL |
| [7] | Goudet J (2001) FSTAT, A Program to Estimate and Test Gene Diversities and Fixation Indices (version 2.9.3). https://www2.unil.ch/popgen/softwares/fstat.htm . (accessed on 2021-08-30) |
| [8] |
Guan B, Peng CC, Wang CH, Jin HZ, Zhang WD (2014) Chemical constituents from the aerial parts of Prinsepia utilis. Chemistry of Natural Compounds, 50, 1106-1107.
DOI URL |
| [9] |
Guan B, Peng CC, Zeng Q, Cheng XR, Yan SK, Jin HZ, Zhang WD (2013) Cytotoxic pentacyclic triterpenoids from Prinsepia utilis. Planta Medica, 79, 365-368.
DOI PMID |
| [10] | He QJ, He JW, Wang YP, Mu YQ, Yang WH, Yang HT, Huang XE, Wu YB, He SL, He JP (2016) Overview of Prinsepia utilis Royle. Chinese Agricultural Science Bulletin, 32(7), 74-78. (in Chinese with English abstract) |
| [ 和琼姬, 和加卫, 王宇萍, 木永青, 杨文宏, 杨洪涛, 黄杏娥, 吴永斌, 和寿莲, 和建平 (2016) 青刺果研究概述. 中国农学通报, 32(7), 74-78.] | |
| [11] |
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965- 1978.
DOI URL |
| [12] |
Huang SQ, Ma YL, Zhang CT, Cai SB, Pang MJ (2017) Bioaccessibility and antioxidant activity of phenolics in native and fermented Prinsepia utilis Royle seed during a simulated gastrointestinal digestion in vitro. Journal of Functional Foods, 37, 354-362.
DOI URL |
| [13] |
Jin WY, Li HW, Wei R, Huang BH, Liu B, Sun TT, Mabberley DJ, Liao PC, Yang Y (2020) New insights into biogeographical disjunctions between Taiwan and the Eastern Himalayas: The case of Prinsepia (Rosaceae). Taxon, 69, 278-289.
DOI URL |
| [14] |
Ju MM, Ma HC, Xin PY, Zhou ZL, Tian B (2015) Development and characterization of EST-SSR markers in Bombax ceiba (Malvaceae). Applications in Plant Sciences, 3, 1500001.
DOI URL |
| [15] |
Kalinowski ST (2005) Hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes, 5, 187-189.
DOI URL |
| [16] |
Li Q, Zhao Y, Xiang XG, Chen JK, Rong J (2019) Genetic diversity of crop wild relatives under threat in Yangtze River basin: Call for enhanced in situ conservation and utilization. Molecular Plant, 12, 1535-1538.
DOI URL |
| [17] | Li XW, Li J (1993) A preliminary floristic study on the seed plants from the region of Hengduan Mountain. Acta Botanica Yunnanica, 15, 217-231. (in Chinese with English abstract) |
| [ 李锡文, 李捷 (1993) 横断山脉地区种子植物区系的初步研究. 云南植物研究, 15, 217-231.] | |
| [18] |
Liu J, Luo YH, Li DZ, Gao LM (2017) Evolution and maintenance mechanisms of plant diversity in the Qinghai- Tibet Plateau and adjacent regions: Restrospect and prospect. Biodiversity Science, 25, 163-174. (in Chinese with English abstract)
DOI |
|
[ 刘杰, 罗亚皇, 李德铢, 高连明 (2017) 青藏高原及毗邻区植物多样性演化与维持机制: 进展及展望. 生物多样性, 25, 163-174.]
DOI |
|
| [19] |
Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ (2013) Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist, 199, 1093-1108.
DOI URL |
| [20] |
Lu LM, Mao LF, Yang T, Ye JF, Liu B, Li HL, Sun M, Miller JT, Mathews S, Hu HH, Niu YT, Peng DX, Chen YH, Smith SA, Chen M, Xiang KL, Le CT, Dang VC, Lu AM, Soltis PS, Soltis DE, Li JH, Chen ZD (2018) Evolutionary history of the angiosperm flora of China. Nature, 554, 234-238.
DOI URL |
| [21] |
Luo D, Yue JP, Sun WG, Xu B, Li ZM, Comes HP, Sun H (2016) Evolutionary history of the subnival flora of the Himalaya‐Hengduan Mountains: First insights from comparative phylogeography of four perennial herbs. Journal of Biogeography, 43, 31-43.
DOI URL |
| [22] |
Ma XG, Wang ZW, Tian B, Sun H (2019) Phylogeographic analyses of the East Asian endemic genus Prinsepia and the role of the East Asian monsoon system in shaping a north-south divergence pattern in China. Frontiers in Genetics, 10, 128.
DOI URL |
| [23] |
Ma YZ, Wang J, Hu QJ, Li JL, Sun YS, Zhang L, Abbott RJ, Liu JQ, Mao KS (2019) Ancient introgression drives adaptation to cooler and drier mountain habitats in a cypress species complex. Communications Biology, 2, 213.
DOI URL |
| [24] |
Muellner-Riehl AN (2019) Mountains as evolutionary arenas: Patterns, emerging approaches, paradigm shifts, and their implications for plant phylogeographic research in the Tibeto-Himalayan region. Frontiers in Plant Science, 10, 195.
DOI PMID |
| [25] |
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-An update. Bioinformatics, 28, 2537-2539.
PMID |
| [26] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
PMID |
| [27] |
Sun H, Zhang JW, Deng T, Boufford DE (2017) Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Diversity, 39, 161-166.
DOI URL |
| [28] |
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535-538.
DOI URL |
| [29] |
Wang S, Shi C, Gao LZ (2013) Plastid genome sequence of a wild woody oil species, Prinsepia utilis, provides insights into evolutionary and mutational patterns of Rosaceae chloroplast genomes. PLoS ONE, 8, e73946.
DOI URL |
| [30] |
Ward FK (1921) The Mekong-Salween Divide as a geographical barrier. The Geographical Journal, 58, 49-56.
DOI URL |
| [31] | Xing YW, Ree RH (2017) Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proceedings of the National Academy of Sciences, USA, 114, E3444-E3451. |
| [32] |
Ye JW, Zhang Y, Wang XJ (2017) Phylogeographic breaks and the mechanisms of their formation in the Sino-Japanese floristic region. Chinese Journal of Plant Ecology, 41, 1003-1019. (in Chinese with English abstract)
DOI URL |
|
[ 叶俊伟, 张阳, 王晓娟 (2017) 中国-日本植物区系中的谱系地理间断及其形成机制. 植物生态学报, 41, 1003-1019.]
DOI |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [11] | Qi Zhang. Suggestions on standardizing the zoological and the botanical nomenclatures of the Himalayas area in the Chinese language [J]. Biodiv Sci, 2023, 31(1): 22131-. |
| [12] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [13] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [14] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [15] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2026 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
