



Biodiv Sci ›› 2020, Vol. 28 ›› Issue (4): 474-484. DOI: 10.17520/biods.2019290 cstr: 32101.14.biods.2019290
• Original Papers: Animal Diversity • Previous Articles Next Articles
Chao Li1,Jinjin Jin1,Jinzhen Luo1,2,Chunhui Wang1,3,Junjie Wang1,Jun Zhao1,*( )
)
Received:2019-09-17
Accepted:2019-12-24
Online:2020-04-20
Published:2020-06-15
Contact:
Jun Zhao
Chao Li,Jinjin Jin,Jinzhen Luo,Chunhui Wang,Junjie Wang,Jun Zhao. Genetic relationships of hatchery populations and wild populations of Tanichthys albonubes near Guangzhou[J]. Biodiv Sci, 2020, 28(4): 474-484.
| 种群 Population | 代号 Code | 所属谱系 Lineage* | 样本量 Sample size* | ||
|---|---|---|---|---|---|
| mtDNA | nuDNA | Microsatellites | |||
| 养殖种群 Hatchery populations | |||||
| 广州芳村 Fangcun, Guangzhou, China | FC | A | 30 | 30 | 30 |
| 新加坡 Singapore | SIG | A | 24 | 24 | 30 |
| 加拿大 Canada | CA | A | 12 | 12 | 12 |
| 野生种群 Wild populations | |||||
| 广州从化良口 Liangkou, Conghua, Guangzhou, China | CL | A | 30 | 30 | 30 |
| 广州从化石门 Shimen, Conghua, Guangzhou, China | SM | A | 30 | 30 | 30 |
| 清远 Qingyuan, China | QY | A | 30 | 30 | 30 |
| 外类群 | |||||
| 深圳 Shenzhen, China | SZ | A | 30 | - | - |
| 总计 Total | 186 | 156 | 162 | ||
Table 1 Sampling information of Tanichthys albonubes of the present study
| 种群 Population | 代号 Code | 所属谱系 Lineage* | 样本量 Sample size* | ||
|---|---|---|---|---|---|
| mtDNA | nuDNA | Microsatellites | |||
| 养殖种群 Hatchery populations | |||||
| 广州芳村 Fangcun, Guangzhou, China | FC | A | 30 | 30 | 30 |
| 新加坡 Singapore | SIG | A | 24 | 24 | 30 |
| 加拿大 Canada | CA | A | 12 | 12 | 12 |
| 野生种群 Wild populations | |||||
| 广州从化良口 Liangkou, Conghua, Guangzhou, China | CL | A | 30 | 30 | 30 |
| 广州从化石门 Shimen, Conghua, Guangzhou, China | SM | A | 30 | 30 | 30 |
| 清远 Qingyuan, China | QY | A | 30 | 30 | 30 |
| 外类群 | |||||
| 深圳 Shenzhen, China | SZ | A | 30 | - | - |
| 总计 Total | 186 | 156 | 162 | ||
| 种群Populations | 芳村 FC | 新加坡SIG | 加拿大CA | 良口 CL | 石门 SM | 清远 QY |
|---|---|---|---|---|---|---|
| 新加坡 SIG | 0.008 | |||||
| 加拿大 CA | 0.009 | 0.001 | ||||
| 良口 CL | 0.006 | 0.006 | 0.007 | |||
| 石门 SM | 0.009 | 0.009 | 0.010 | 0.007 | ||
| 清远 QY | 0.007 | 0.001 | 0.002 | 0.005 | 0.008 | |
| 深圳 SZ | 0.013 | 0.014 | 0.015 | 0.012 | 0.015 | 0.014 |
Table 2 K2P genetic distance among seven populations of Tanichthys albonubes based on Cyt b gene. Population codes refer to Table 1.
| 种群Populations | 芳村 FC | 新加坡SIG | 加拿大CA | 良口 CL | 石门 SM | 清远 QY |
|---|---|---|---|---|---|---|
| 新加坡 SIG | 0.008 | |||||
| 加拿大 CA | 0.009 | 0.001 | ||||
| 良口 CL | 0.006 | 0.006 | 0.007 | |||
| 石门 SM | 0.009 | 0.009 | 0.010 | 0.007 | ||
| 清远 QY | 0.007 | 0.001 | 0.002 | 0.005 | 0.008 | |
| 深圳 SZ | 0.013 | 0.014 | 0.015 | 0.012 | 0.015 | 0.014 |
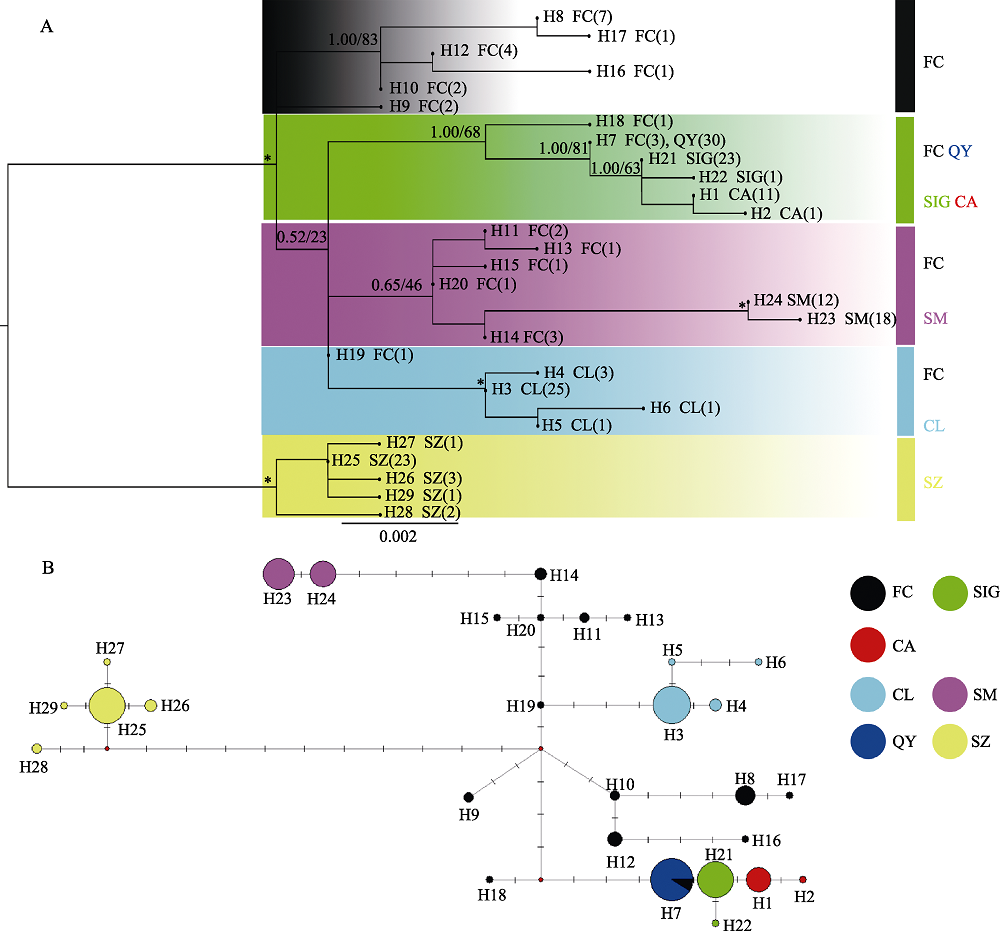
Fig. 2 Phylogenetic trees and haplotype networks of Tanichthys albonubes based on Cyt b gene. Population codes refer to Table 1. (A) Bayesian inference (BI) and maximum-likelihood (ML) trees of T. albonubes based on Cyt b gene. The asterisks on the branches indicate posterior probabilities (PP) ≥ 0.90 and bootstrap values (BS) ≥ 90. (B) Haplotype networks of T. albonubes based on Cyt b gene (segments above lines indicate the number of mutations).
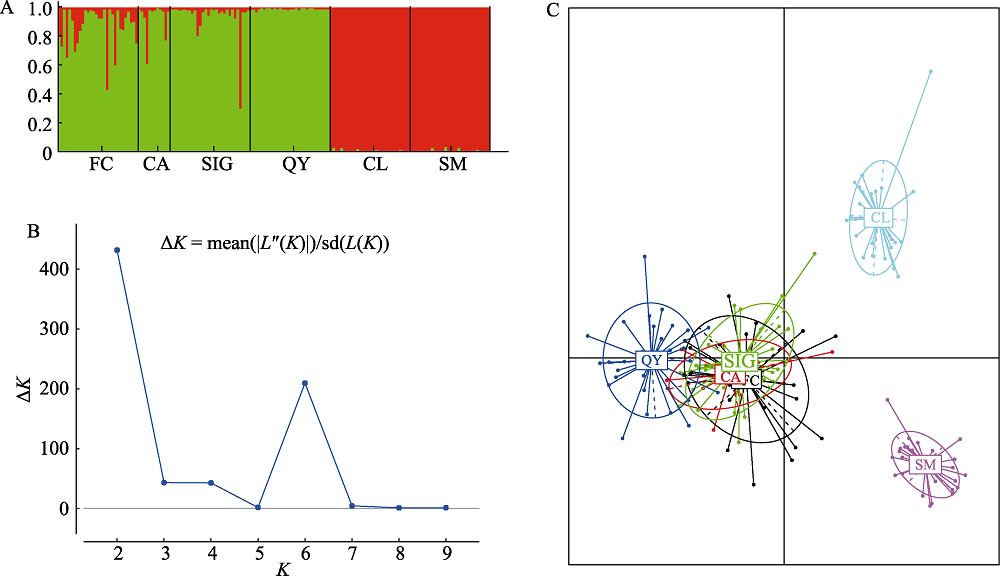
Fig. 3 Population structure analysis of hatchery and wild populations of Tanichthys albonubes. (A) STRUCTURE plots of K = 2 based on 13 microsatellite loci; (B) ΔK values of the different number of clusters (K); (C) PCA analysis based on 13 microsatellite loci. Population codes refer to Table 1.
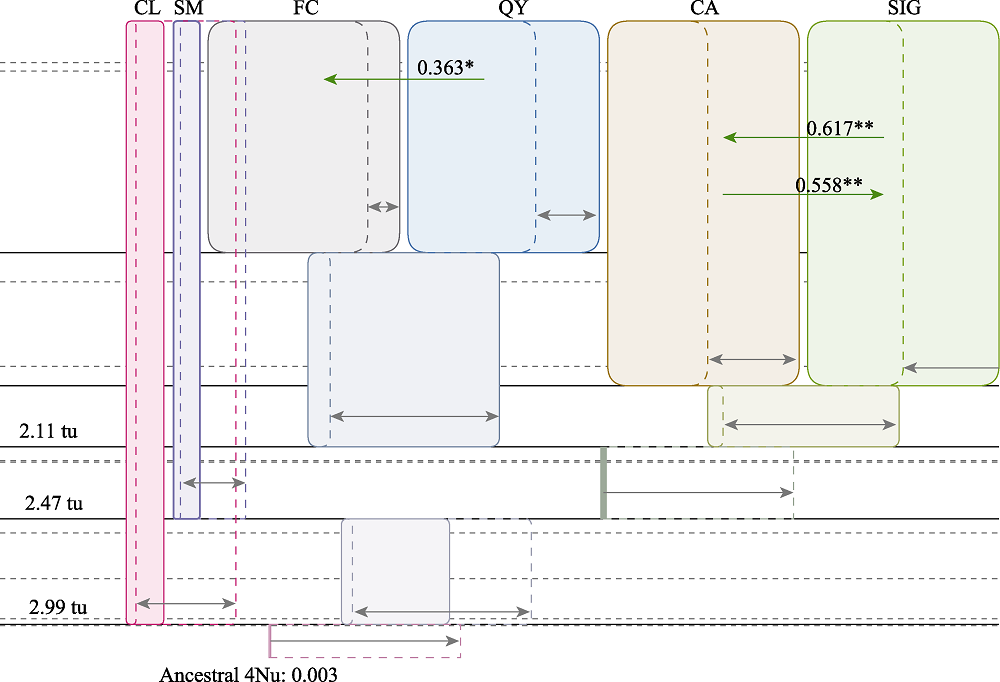
Fig. 4 Gene flow estimation among hatchery and wild populations of Tanichthys albonubes. Grey arrows and boxes were marginal distribution values in demographic units. tu means time since ancestral population splitting. Green arrows and the numbers above them showed the gene flows (2Nm values) and their directions. Only statistically significant cases of gene flow are presented. * P < 0.05, ** P < 0.01. Population codes refer to Table 1.
| [1] |
Bandelt HJ, Forster P, RŏHL A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
DOI URL |
| [2] |
Cao L, Zhang E, Zang CX, Cao WX (2016) Evaluating the status of China’s continental fish and analyzing their causes of endangerment through the red list assessment. Biodiversity Science, 24, 598-609. (in Chinese with English abstract)
DOI URL |
|
[ 曹亮, 张鹗, 臧春鑫, 曹文宣 (2016) 通过红色名录评估研究中国内陆鱼类受威胁现状及其成因. 生物多样性, 24, 598-609.]
DOI URL |
|
| [3] |
Chan B, Chen X (2009) Discovery of Tanichthys albonubes Lin 1932 (Cyprinidae) on Hainan Island and notes on its ecology. Zoological Research, 30, 209-214.
DOI URL |
| [4] | Chen GZ, Fang ZQ, Ma GZ (2004) Embryonic development of Tanichthys albonubes. Journal of Fishery Sciences of China, 11, 489-496. (in Chinese with English abstract) |
| [ 陈国柱, 方展强, 马广智 (2004) 唐鱼胚胎发育观察. 中国水产科学, 11, 489-496.] | |
| [5] | Chen YY (1998) Fauna Sinica • Osteichthyes : Cypriniformes (II). Science Press, Beijing. (in Chinese) |
| [ 陈宜瑜 (1998) 中国动物志•硬骨鱼纲: 鲤形目(中卷). 科学出版社, 北京.] | |
| [6] |
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359-361.
DOI URL |
| [7] | Eastman OR (1938) Tanichthys albonubes Lin, the new wonder fish. The Aquarium, 7, 73-75. |
| [8] |
Ellstrand NC, Rieseberg LH (2016) When gene flow really matters: Gene flow in applied evolutionary biology. Evolutionary Applications, 9, 833-836.
DOI URL |
| [9] |
Evanno GS, Regnaut SJ, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611-2620.
DOI URL |
| [10] | Freyhof J, Herder F (2001) Tanichthys micagemmae, a new miniature cyprinid fish from Central Vietnam (Cypriniformes: Cyprinidae). Ichthyological Exploration of Freshwaters, 12, 215-220. |
| [11] |
Glover KA, Solberg MF, McGinnity P, Hindar K, Verspoor E, Coulson MW, Svåsand T (2017) Half a century of genetic interaction between farmed and wild Atlantic salmon: Status of knowledge and unanswered questions. Fish and Fisheries, 18, 890-927.
DOI URL |
| [12] |
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307-321.
DOI URL |
| [13] | Hey J, Chung Y, Sethuraman A, Lachance J, Tishkoff S, Sousa VC (2018) Phylogeny estimation by integration over isolation with migration models. Molecular Biology and Evolution, 35, 2805-2818. |
| [14] |
Jaisuk C, Senanan W (2018) Effects of landscape features on population genetic variation of a tropical stream fish, stone lapping minnow, Garra cambodgiensis, in the upper Nan River drainage basin, northern Thailand. PeerJ, 6, e4487.
DOI URL |
| [15] |
Jombart T (2008) adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403-1405.
DOI URL |
| [16] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547-1549.
DOI URL |
| [17] |
Larson G, Burger J (2013) A population genetics view of animal domestication. Trends in Genetics, 29, 197-205.
DOI URL |
| [18] |
Lefort V, Longueville JE, Gascuel O (2017) SMS: Smart model selection in PhyML. Molecular Biology and Evolution, 34, 2422-2424.
DOI URL |
| [19] | Li J, Li XH (2011) A new record of fish Tanichthys albonubes (Cypriniformes: Cyprinidae) in Guangxi, China. Chinese Journal of Zoology, 46, 136-140. (in Chinese) |
| [ 李捷, 李新辉 (2011) 广西鱼类一新纪录: 唐鱼(鲤形目: 鲤科). 动物学杂志, 46, 136-140.] | |
| [20] |
Librado P, Rozas J (2009) DNASP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI URL |
| [21] | Lin SY (1932) New cyprinid fishes from White Cloud Mountain, Canton. Lingnan Science Journal, 11, 379-383. |
| [22] | Liu HS, Yi ZS, Liang JH, Li WH, Lin XT (2008) Morphological variations between the wild population and hatchery stock of Tanichtys albonubes. Journal of Jinan University, 29, 295-299. (in Chinese with English abstract) |
| [ 刘汉生, 易祖盛, 梁建宏, 李伟华, 林小涛 (2008) 唐鱼野生种群和养殖群体的形态差异分析. 暨南大学学报, 29, 295-299.] | |
| [23] |
Luo JZ, Lin HD, Yang F, Yi ZS, Chan BP, Zhao J (2015) Population genetic structure in wild and hatchery populations of white cloud mountain minnow (Tanichthys albonubes): Recommendations for conservation. Biochemical Systematics and Ecology, 62, 142-150.
DOI URL |
| [24] | Pan JH (1990) Freshwater Fishes of Gongdong Province. Guangdong Science and Technology Press, Guangzhou. (in Chinese) |
| [ 潘炯华 (1990) 广东淡水鱼类志. 广东科技出版社, 广州.] | |
| [25] | Pritchard JK, Stephens P, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959. |
| [26] | R Development Core Team (2015) R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing, Vienna. |
| [27] |
Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard M, Huelsenbeck J (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542.
DOI URL |
| [28] |
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312-1313.
DOI URL |
| [29] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins, DG (1997) The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI URL |
| [30] | Weitzman SH, Chan LL (1966) Identification and relationships of Tanichthys albonubes and Aphyocypris pooni, two cyprinid fishes from South China and Hong Kong. Copeia, 1966, 285-296. |
| [31] |
Yan F, Lv JC, Zhang BL, Yuan ZY, Zhao HP, Huang S, Wei G, Mi X, Zou DH, Xu W, Chen S, Wang J, Xie F, Wu MY, Xiao HB, Liang ZQ, Jin JQ, Wu SF, Xu CS, Tapley B, Turvey ST, Papenfuss TJ, Cunningham AA, Murphy RW, Zhang YP, Che J (2018) The Chinese giant salamander exemplifies the hidden extinction of cryptic species. Current Biology, 28, R590-R592.
DOI URL |
| [32] | Yi ZS, Chen XL, Wu JX, Yu SC, Huang CE (2004) Rediscovering the wild population of White Cloud Mountain minnows (Tanichthys albonubes Lin) in Guangdong Province. Zoological Research, 25, 551-555. (in Chinese) |
| [ 易祖盛, 陈湘粦, 巫锦雄, 余顺昌, 黄慈恩 (2004) 野生唐鱼在广东的再发现. 动物学研究, 25, 551-555.] | |
| [33] | Zhang XX, Zhu QY, Zhao, J (2017) Geometric morphometric analysis of body-form variability in populations of Tanichthys albonubes. Journal of Fisheries of China, 41, 1365-1372. (in Chinese with English abstract) |
| [ 张秀霞, 朱巧莹, 赵俊 (2017) 利用几何形态测量学方法分析唐鱼群体的形态变异. 水产学报, 41, 1365-1373.] | |
| [34] | Zhao J, Hsu CK, Luo JZ, Wang CH, Chan BP, Li J, Kuo PH, Lin HD (2018) Genetic diversity and population history of Tanichthys albonubes (Teleostei: Cyprinidae): Implications for conservation. Aquatic Conversation: Marine and Freshwater Ecosystem, 28, 422-434. |
| [35] | Zhao J, Yi ZS, Zhou XY, Xiao Z (2010) Background Resources of Aquatic Plants and Animals in Guangzhou. Science Press, Beijing. (in Chinese) |
| [ 赵俊, 易祖盛, 周叶先, 肖智 (2010) 广州市水生动植物本底资源. 科学出版社, 北京.] | |
| [36] | Zheng CY (1989) The Fishes of the Zhujiang River. Science Press, Beijing. (in Chinese) |
| [ 郑慈英 (1989) 珠江鱼类志. 科学出版社, 北京.] |
| [1] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [2] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [3] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [4] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [5] | Qiong Sun, Rong Wang, Xiaoyong Chen. Genomic island of divergence during speciation and its underlying mechanisms [J]. Biodiv Sci, 2022, 30(3): 21383-. |
| [6] | Junwei Ye, Bin Tian. Genetic structure and its causes of an important woody oil plant in Southwest China, Prinsepia utilis (Rosaceae) [J]. Biodiv Sci, 2021, 29(12): 1629-1637. |
| [7] | Xunhe Huang, Zheqi Yu, Zhuoxian Weng, Danlin He, Zhenhua Yi, Weina Li, Jiebo Chen, Xiquan Zhang, Bingwang Du, Fusheng Zhong. Mitochondrial genetic diversity and maternal origin of Guangdong indigenous chickens [J]. Biodiv Sci, 2018, 26(3): 238-247. |
| [8] | Jiping Yang, Ce Li, Weitao Chen, Yuefei Li, Xinhui Li. Genetic diversity and population demographic history of Ochetobius elongatus in the middle and lower reaches of the Xijiang River [J]. Biodiv Sci, 2018, 26(12): 1289-1295. |
| [9] | Jian-Feng Mao, Yongpeng Ma, Renchao Zhou. Approaches used to detect and test hybridization: combining phylogenetic and population genetic analyses [J]. Biodiv Sci, 2017, 25(6): 577-599. |
| [10] | Qiujie Zhou, Yacheng Cai, Wei Lun Ng, Wei Wu, Seping Dai, Feng Wang, Renchao Zhou. Molecular evidence for natural hybridization between two Melastoma species endemic to Hainan and their widespread congeners [J]. Biodiv Sci, 2017, 25(6): 638-646. |
| [11] | Jianhua Xue, Li Jiang, Xiaolin Ma, Yanhong Bing, Sichen Zhao, Keping Ma. Identification of lotus cultivars using DNA fingerprinting [J]. Biodiv Sci, 2016, 24(1): 3-11. |
| [12] | Haomin Lyu, Renchao Zhou, Suhua Shi. Recent advances in the study of ecological speciation [J]. Biodiv Sci, 2015, 23(3): 398-407. |
| [13] | Changhuan He, Yu Zhou, Lifan Wang, Li Zhang. Estimating population size and genetic diversity of Asian elephant in the Shangyong Nature Reserve [J]. Biodiv Sci, 2015, 23(2): 202-209. |
| [14] | Min Xiong, Shuang Tian, Zhirong Zhang, Dengmei Fan, Zhiyong Zhang. Population genetic structure and conservation units of Sinomanglietia glauca (Magnoliaceae) [J]. Biodiv Sci, 2014, 22(4): 476-484. |
| [15] | Wei Zhou,Hong Wang. Pollen dispersal analysis using DNA markers [J]. Biodiv Sci, 2014, 22(1): 97-108. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn