



Biodiv Sci ›› 2016, Vol. 24 ›› Issue (11): 1296-1305. DOI: 10.17520/biods.2016202 cstr: 32101.14.biods.2016202
Special Issue: 昆虫多样性与生态功能
• Original Papers: Animal Diversity • Previous Articles Next Articles
Qian Jin1,2, Fen Chen1, Guijie Luo1, Weijia Cai1, Xu Liu1, Hao Wang1, Caiqing Yang2, Mengdi Hao2, Aibing Zhang2,*( )
)
Received:2016-07-22
Accepted:2016-11-02
Online:2016-11-20
Published:2016-12-14
Contact:
Zhang Aibing
Qian Jin, Fen Chen, Guijie Luo, Weijia Cai, Xu Liu, Hao Wang, Caiqing Yang, Mengdi Hao, Aibing Zhang. Estimation of species richness of moths (Insecta: Lepidoptera) based on DNA barcoding in Suqian, China[J]. Biodiv Sci, 2016, 24(11): 1296-1305.
| MOTU① | 种名 Species② | MOTU | 种名 Species |
|---|---|---|---|
| MOTU01 | Lacanobia contigua | MOTU34 | Parapoynx vittalis |
| MOTU02 | Agrotis segetum 黄地老虎 | MOTU35 | Heliothis assulta 烟青虫 |
| MOTU03 | Conogethes punctiferalis 桃蛀螟 | MOTU36 | Herminia grisealis |
| MOTU04 | Nola cicatricalis | MOTU37 | Spodoptera depravata |
| MOTU05 | Hipoepa fractalis 中影单跗夜蛾 | MOTU38 | Scopula subpunctaria |
| MOTU06 | Oraesia lata 平嘴壶夜蛾 | MOTU39 | Diaphania indica 瓜绢野螟 |
| MOTU07 | Spodoptera litura 斜纹夜蛾 | MOTU40 | Botyodes diniasalis 黄翅缀叶野螟 |
| MOTU08 | Uropyia meticulodina 核桃美舟蛾 | MOTU41 | Emmelia trabealis 谐夜蛾 |
| MOTU09 | Evergestis extimalis | MOTU42 | Termioptycha nigrescens |
| MOTU10 | Mythimna separate | MOTU43 | Ipimorpha subtusa 杨逸色夜蛾 |
| MOTU11 | Ctenoplusia albostriata | MOTU44 | Spilosoma lubricipeda |
| MOTU12 | Peridea lativitta 侧带内斑舟蛾 | MOTU45 | Parapediasia teterrellus |
| MOTU13 | Thyas juno 肖毛翅夜蛾 | MOTU46 | Adoxophyes orana |
| MOTU14 | Nephopterix fumella | MOTU47 | Simplicia rectalis 黑点贫夜蛾 |
| MOTU15 | Spodoptera exigua 甜菜夜蛾 | MOTU48 | Glyptoteles leucacrinella 亮雕斑螟 |
| MOTU16 | Oglasa consanguis | MOTU49 | Somena scintillans |
| MOTU17 | Diaphania perspectalis 黄杨绢野螟 | MOTU50 | Anadevidia peponis 葫芦夜蛾 |
| MOTU18 | Athetis lepigone | MOTU51 | Hypocala subsatura 苹梢鹰夜蛾 |
| MOTU19 | Miyakea raddeella | MOTU52 | Choristoneura diversana 异色卷蛾 |
| MOTU20 | Herpetogramma pseudomagna 狭翅切叶野螟 | MOTU53 | Choristoneura luticostana 棕色卷蛾 |
| MOTU21 | Axylia putris 朽木夜蛾 | MOTU54 | Archips betulana; Archips podana |
| MOTU22 | Mocis ancilla | MOTU55 | Palpita nigropunctalis 白蜡绢须野螟 |
| MOTU23 | Mecyna tricolor | MOTU56 | Epiblema foenella 白钩小卷蛾 |
| MOTU24 | Mamestra brassicae 甘蓝夜蛾 | MOTU57 | Grammodes geometrica 象夜蛾 |
| MOTU25 | Xanthorhoe quadrifasiata | MOTU58; MOTU60 | Plusia nadeja |
| MOTU26 | Euproctis similis | MOTU59 | Sphinx caligineus |
| MOTU27 | Zanclognatha lunalis 朽镰须夜蛾 | MOTU61 | Acronicta rumicis 梨剑纹夜蛾 |
| MOTU28 | Theretra japonica | MOTU62 | Herminia tarsipennalis |
| MOTU29 | Herminia tarsicrinalis | MOTU63 | Pangrapta trimantesalis |
| MOTU30 | Eremodrina morosa | MOTU64 | Eutelia hamulatrix 沟尾夜蛾 |
| MOTU31 | Helicoverpa armigera 棉铃虫 | MOTU65 | Plusilla rosalia |
| MOTU32 | Nomophila noctuella | MOTU66 | Bertula bistrigata |
| MOTU33 | Dysgonia mandschuriana | MOTU67 | Gastropacha populifolia 杨枯叶蛾 |
Table 1 The comparison of ABGD method with morphospecies
| MOTU① | 种名 Species② | MOTU | 种名 Species |
|---|---|---|---|
| MOTU01 | Lacanobia contigua | MOTU34 | Parapoynx vittalis |
| MOTU02 | Agrotis segetum 黄地老虎 | MOTU35 | Heliothis assulta 烟青虫 |
| MOTU03 | Conogethes punctiferalis 桃蛀螟 | MOTU36 | Herminia grisealis |
| MOTU04 | Nola cicatricalis | MOTU37 | Spodoptera depravata |
| MOTU05 | Hipoepa fractalis 中影单跗夜蛾 | MOTU38 | Scopula subpunctaria |
| MOTU06 | Oraesia lata 平嘴壶夜蛾 | MOTU39 | Diaphania indica 瓜绢野螟 |
| MOTU07 | Spodoptera litura 斜纹夜蛾 | MOTU40 | Botyodes diniasalis 黄翅缀叶野螟 |
| MOTU08 | Uropyia meticulodina 核桃美舟蛾 | MOTU41 | Emmelia trabealis 谐夜蛾 |
| MOTU09 | Evergestis extimalis | MOTU42 | Termioptycha nigrescens |
| MOTU10 | Mythimna separate | MOTU43 | Ipimorpha subtusa 杨逸色夜蛾 |
| MOTU11 | Ctenoplusia albostriata | MOTU44 | Spilosoma lubricipeda |
| MOTU12 | Peridea lativitta 侧带内斑舟蛾 | MOTU45 | Parapediasia teterrellus |
| MOTU13 | Thyas juno 肖毛翅夜蛾 | MOTU46 | Adoxophyes orana |
| MOTU14 | Nephopterix fumella | MOTU47 | Simplicia rectalis 黑点贫夜蛾 |
| MOTU15 | Spodoptera exigua 甜菜夜蛾 | MOTU48 | Glyptoteles leucacrinella 亮雕斑螟 |
| MOTU16 | Oglasa consanguis | MOTU49 | Somena scintillans |
| MOTU17 | Diaphania perspectalis 黄杨绢野螟 | MOTU50 | Anadevidia peponis 葫芦夜蛾 |
| MOTU18 | Athetis lepigone | MOTU51 | Hypocala subsatura 苹梢鹰夜蛾 |
| MOTU19 | Miyakea raddeella | MOTU52 | Choristoneura diversana 异色卷蛾 |
| MOTU20 | Herpetogramma pseudomagna 狭翅切叶野螟 | MOTU53 | Choristoneura luticostana 棕色卷蛾 |
| MOTU21 | Axylia putris 朽木夜蛾 | MOTU54 | Archips betulana; Archips podana |
| MOTU22 | Mocis ancilla | MOTU55 | Palpita nigropunctalis 白蜡绢须野螟 |
| MOTU23 | Mecyna tricolor | MOTU56 | Epiblema foenella 白钩小卷蛾 |
| MOTU24 | Mamestra brassicae 甘蓝夜蛾 | MOTU57 | Grammodes geometrica 象夜蛾 |
| MOTU25 | Xanthorhoe quadrifasiata | MOTU58; MOTU60 | Plusia nadeja |
| MOTU26 | Euproctis similis | MOTU59 | Sphinx caligineus |
| MOTU27 | Zanclognatha lunalis 朽镰须夜蛾 | MOTU61 | Acronicta rumicis 梨剑纹夜蛾 |
| MOTU28 | Theretra japonica | MOTU62 | Herminia tarsipennalis |
| MOTU29 | Herminia tarsicrinalis | MOTU63 | Pangrapta trimantesalis |
| MOTU30 | Eremodrina morosa | MOTU64 | Eutelia hamulatrix 沟尾夜蛾 |
| MOTU31 | Helicoverpa armigera 棉铃虫 | MOTU65 | Plusilla rosalia |
| MOTU32 | Nomophila noctuella | MOTU66 | Bertula bistrigata |
| MOTU33 | Dysgonia mandschuriana | MOTU67 | Gastropacha populifolia 杨枯叶蛾 |
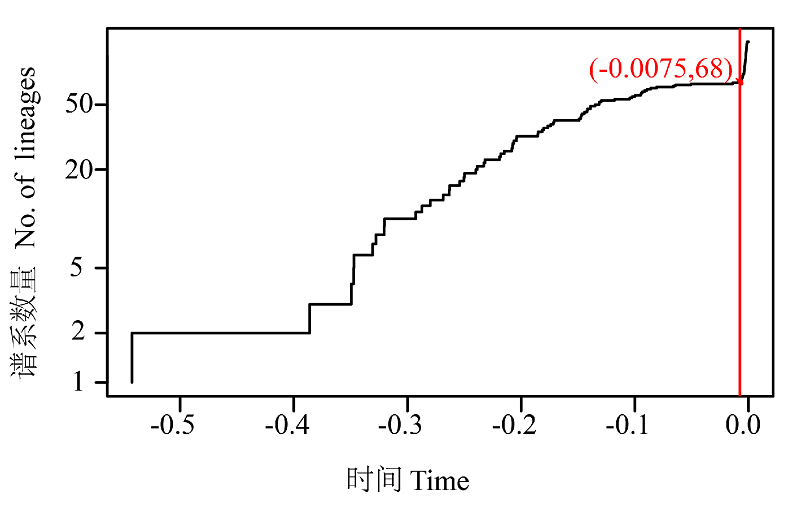
Fig. 2 Lineages-through-time plot based on the time calibrated tree obtained from all 121 haplotypes. The sharp increase in branching rate, corresponding to the transition from interspecies to intraspecies branching events.
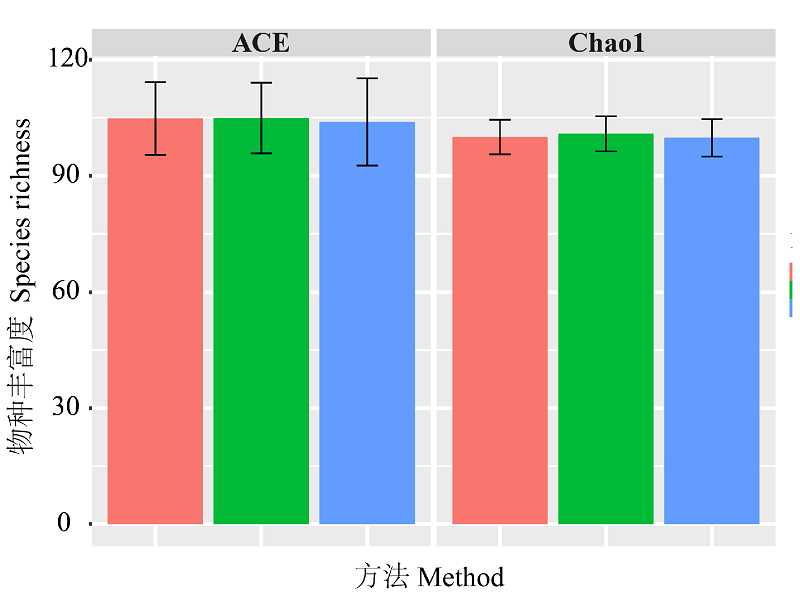
Fig. 3 Species richness estimation results with different methods. Pink colour represents ABGD-based method. Green colour represents GMYC-based method. Blue colour represents morphology-based method. Black error bar indicates confidence interval.
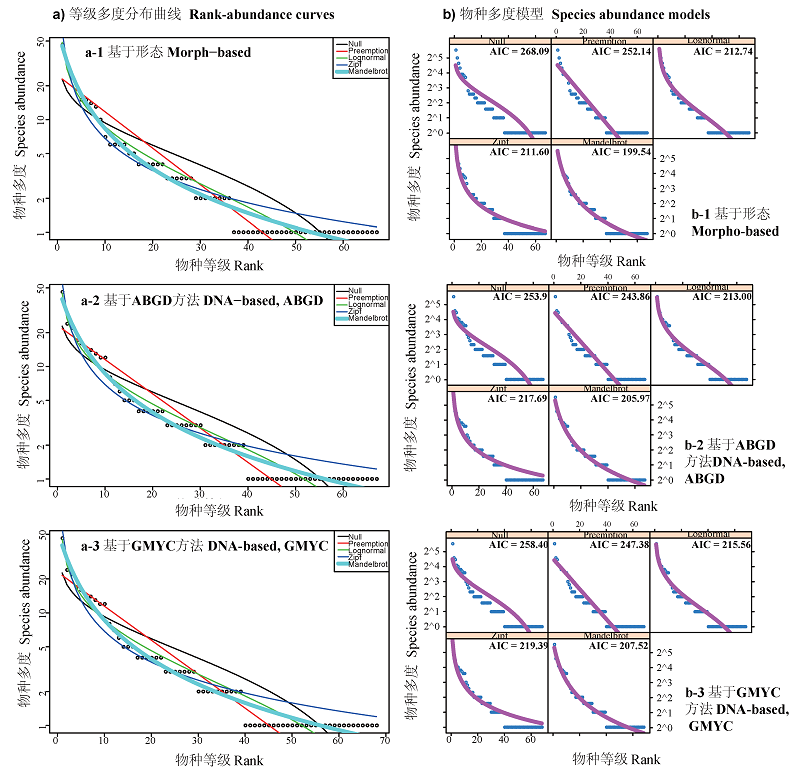
Fig. 4 Rank-abundance curves and fittings of five models. a-1)Species rank-abundance curves based on morphological method, a-2) Species rank-abundance curves based on ABGD method, a-3) Species rank-abundance curves based on GMYC method; b-1) Species abundance models based on morphological method, b-2) Species abundance models based on ABGD method, b-3) Species abundance models based on GMYC method. Black points represent the real abundance data. Black curve represents broken stick model. Red curve represents preemption model. Green curve represents log normal model. Dark blue curve represents Zipf model. Light blue curve represents Zipf-Mandelbrot model.
| 1 | Austerlitz F, David O, Schaeffer B, Bleakley K, Olteanu M, Leblois R, Veuille M, Laredo C (2009) DNA barcode analysis: a comparison of phylogenetic and statistical classification methods. BMC Bioinformatics, 10(Suppl. 14), 1-13. |
| 2 | Blaxter M, Floyd R (2003) Molecular taxonomics for biodiversity surveys: already a reality. Trends in Ecology and Evolution, 18, 268-269. |
| 3 | Boykin LM, Armstrong KF, Kubatko L, Barro PD (2012) Species delimitation and global biosecurity. Evolutionary Bioinformatics, 8, 1-37. |
| 4 | Burnham KP, Anderson DR (2002) Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer-Verlag, New York. |
| 5 | Chao A (1984) Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics, 11, 265-270. |
| 6 | Chao A, Hwang WH, Chen YC, Kuo CY (2000) Estimating the number of shared species in two communities. Statistica Sinica, 10, 227-246. |
| 7 | Colwell RK, Coddington JA (1994) Estimating terrestrial biodiversity through extrapolation. Philosophical Transactions of the Royal Society B: Biological Sciences, 345, 101-118. |
| 8 | Darriba D, Taboada GL, Doallo R, Posada D (2012) jModel- Test 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. |
| 9 | Decaëns T, Porco D, James SW, Brown G, Da SE, Dupont L, Lapied E, Rougerie R, Taberlet P, Roy V (2015) Dissecting tropical earthworm biodiversity patterns in tropical rainforests through the use of DNA barcoding. Genome, 58, 210. |
| 10 | Dincă V, Zakharov EV, Hebert PDN, Vila R (2011) Complete DNA barcode reference library for a country’s butterfly fauna reveals high performance for temperate Europe. Proceedings of the Royal Society B: Biological Sciences, 278, 347-355. |
| 11 | Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology, 4, e88. |
| 12 | Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214. |
| 13 | Ebach MC, Holdrege C (2005) DNA barcoding is no substitute for taxonomy. Nature, 434, 697. |
| 14 | Ekrem T, Willassen E, Stur E (2007) A comprehensive DNA sequence library is essential for identification with DNA barcodes. Molecular Phylogenetics and Evolution, 43, 530-542. |
| 15 | Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Systematic Biology, 62, 707-724. |
| 16 | Gomez-Alvarez V, King GM, Nüsslein K (2007) Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiology Ecology, 60, 60-73. |
| 17 | Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270, 313-321. |
| 18 | Hebert PDN, Humble LM (2011) A comprehensive DNA barcode library for the looper moths (Lepidoptera: Geometridae) of British Columbia, Canada. PLoS ONE, 6, e18290. |
| 19 | Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences, USA, 101, 14812-14817. |
| 20 | Herrera A, Héry M, Stach JEM, Jaffré T, Normand P, Navarro E (2007) Species richness and phylogenetic diversity comparisons of soil microbial communities affected by nickel-mining and revegetation efforts in New Caledonia. European Journal of Soil Biology, 43, 130-139. |
| 21 | Hickerson MJ, Meyer CP, Moritz C (2006) DNA barcoding will often fail to discover new animal species over broad parameter space. Systematic Biology, 55, 729-739. |
| 22 | Jin Q, He LJ, Zhang AB (2012) A simple 2D non-parametric resampling statistical approach to assess confidence in species identification in DNA barcoding—an alternative to Likelihood and Bayesian approaches. PLoS ONE, 7, e50831. |
| 23 | Jin Q, Han HL, Hu XM, Li XH, Zhu CD, Ho SYW, Ward RD, Zhang AB (2013) Quantifying species diversity with a DNA barcoding-based method: Tibetan moth species (Noctuidae) on the Qinghai-Tibetan Plateau. PLoS ONE, 8, e64428. |
| 24 | Jin Q, Zhang AB (2013) Distance-based DNA barcoding methods for insects. Chinese Journal of Applied Entomology, 50, 283-287. (in Chinese) |
| [金倩, 张爱兵 (2013) 昆虫DNA条形码分析中的距离方法. 应用昆虫学报, 50, 283-287.] | |
| 25 | Kempton RA, Taylor LR (1974) Log-series and log-normal parameters as diversity discriminants for the Lepidoptera. The Journal of Animal Ecology, 43, 381-399. |
| 26 | Leray M, Knowlton N (2015) DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proceedings of the National Academy of Sciences, USA, 112, 2076-2081. |
| 27 | Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2015) Plant DNA barcoding: from gene to genome. Biological Reviews, 90, 157-166. |
| 28 | MacArthur RH (1957) On the relative abundance of bird species. Proceedings of the National Academy of Sciences, USA, 43, 293-295. |
| 29 | Mandelbrot BB (1983) The Fractal Geometry of Nature. W.H. Freeman and Company, San Francisco. |
| 30 | May RM (1988) How many species are there on earth? Science, 241, 1441-1449. |
| 31 | Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biology, 3, e422. |
| 32 | O’hara RB (2005) Species richness estimators: how many species can dance on the head of a pin? Journal of Animal Ecology, 74, 375-386. |
| 33 | Oline DK (2006) Phylogenetic comparisons of bacterial communities from serpentine and nonserpentine soils. Applied and Environmental Microbiology, 72, 6965-6971. |
| 34 | Ovreas L (2000) Population and community level approaches for analyzing microbial diversity in natural environments. Ecology Letters, 3, 236-251. |
| 35 | Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology, 55, 595-609. |
| 36 | Preston FW (1948) The commonness, and rarity, of species. Ecology, 29, 254-283. |
| 37 | Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology, 21, 1864-1877. |
| 38 | Qin J, Zhang YZ, Zhou X, Kong XB, Wei SJ, Ward RD, Zhang AB (2015) Mitochondrial phylogenomics and genetic relationships of closely related pine moth (Lasiocampidae: Dendrolimus) species in China, using whole mitochondrial genomes. BMC Genomics, 16, 428. |
| 39 | Quicke DLJ, Smith MA, Janzen DH, Hallwachs W, Fernandez-Triana J, Laurenne NM, Zaldivar-Riveron A, Shaw MR, Broad GR, Klopfstein S, Shaw SR, Hrcek J, Hebert PDN, Miller SE, Rodriguez JJ, Whitfield JB, Sharkey MJ, Sharanowski BJ, Jussila R, Gauld ID, Chesters D, Vogler AP (2012) Utility of the DNA barcoding gene fragment for parasitic wasp phylogeny (Hymenoptera: Ichneumonoidea): data release and new measure of taxonomic congruence. Molecular Ecology Resources, 12, 676-685. |
| 40 | Rambaut A, Drummond AJ (2007) Tracer v1.4: MCMC Trace Analyses Tool. . |
| 41 | Ratnasingham S, Hebert PD (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE, 8, e66213. |
| 42 | Rubinoff D (2006) DNA barcoding evolves into the familiar. Conservation Biology, 20, 1548-1549. |
| 43 | Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425. |
| 44 | Schindel DE, Miller SE (2005) DNA barcoding a useful tool for taxonomists. Nature, 435, 17. |
| 45 | Smith MA, Fisher BL, Hebert PDN (2005) DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Philosophical Transactions of the Royal Society B: Biological Sciences, 360, 1825-1834. |
| 46 | Tokeshi M (1990) Niche apportionment or random assortment: species abundance patterns revisited. The Journal of Animal Ecology, 10, 1129-1146. |
| 47 | Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294-299. |
| 48 | Ward RD, Hanner R, Hebert PDN (2009) The campaign to DNA barcode all fishes, FISH-BOL. Journal of Fish Biology, 74, 329-356. |
| 49 | Wiemers M, Fiedler K (2007) Does the DNA barcoding gap exist? — a case study in blue butterflies (Lepidoptera: Lycaenidae). Frontiers in Zoology, 4, 8. |
| 50 | Wilson JB (1991) Methods for fitting dominance/diversity curves. Journal of Vegetation Science, 2, 35-46. |
| 51 | Zhang AB, Feng J, Ward RD, Wan P, Gao Q, Wu J, Zhao WZ (2012a) A new method for species identification via protein-coding and non-coding DNA barcodes by combining machine learning with bioinformatic methods. PLoS ONE, 7, e30986. |
| 52 | Zhang AB, Muster C, Liang HB, Zhu CD, Crozier R, Wan P, Feng J, Ward RD (2012b) A fuzzy-set-theory-based approach to analyse species membership in DNA barcoding. Molecular Ecology, 21, 1848-1863. |
| 53 | Zhang AB, Sikes DS, Muster C, Li SQ (2008) Inferring species membership using DNA sequences with back-propagation neural networks. Systematic Biology, 57, 202-215. |
| 54 | Zipf GK (1949) Human Behavior and the Principle of Least Effort. Hafner, New York. |
| [1] | Jia Zhenni, Zhang Yicen, Du Yanjun, Ren Haibao. Influences of disturbances on successional dynamics of species diversity in mid- subtropical forests [J]. Biodiv Sci, 2025, 33(2): 24078-. |
| [2] | Xinyi He, Yumei Pan, Yan Zhu, Jiayi Chen, Sirong Zhang, Naili Zhang. Impact of ectomycorrhizal tree dominance and species richness on soil nitrogen turnover in a warm temperate forest [J]. Biodiv Sci, 2024, 32(9): 24173-. |
| [3] | Yiyun Gu, Jiaqi Xue, Jinhui Gao, Xinyi Xie, Ming Wei, Jinyu Lei, Cheng Wen. A public science data-based regional bird diversity assessment method [J]. Biodiv Sci, 2024, 32(7): 24080-. |
| [4] | Jingci Meng, Guodong Wang, Guanglan Cao, Nanlin Hu, Meiling Zhao, Yantong Zhao, Zhenshan Xue, Bo Liu, Wenhua Piao, Ming Jiang. Patterns and drivers of plant species richness in Phragmites australis marshes in China [J]. Biodiv Sci, 2024, 32(2): 23194-. |
| [5] | Guoshan Shi, Feng Liu, Guanghong Cao, Dian Chen, Shangwen Xia, Yun Deng, Bin Wang, Xiaodong Yang, Luxiang Lin. Beta diversity of woody plants in a tropical seasonal rainforest at Xishuangbanna: Roles of space, environment, and forest stand structure [J]. Biodiv Sci, 2024, 32(12): 24285-. |
| [6] | Nuriye Muhetaier, Xiuying Zhang, Subinuer Eli, Houhun Li. Annual report of new taxa for Chinese Lepidoptera in 2023 [J]. Biodiv Sci, 2024, 32(11): 24428-. |
| [7] | Liyuan Wang, Huijian Hu, Jie Jiang, Yiming Hu. Species richness patterns of mammals and birds and their drivers in the Nanling Mountain Range [J]. Biodiv Sci, 2024, 32(1): 23026-. |
| [8] | Zhifa Liu, Xincai Wang, Yuening Gong, Daojian Chen, Qiang Zhang. Diversity and elevational distribution of birds and mammals based on infrared camera monitoring in Guangdong Nanling National Nature Reserve [J]. Biodiv Sci, 2023, 31(8): 22689-. |
| [9] | Chao Xing, Yi Lin, Zhiqiang Zhou, Lianjun Zhao, Shiwei Jiang, Zhenzhen Lin, Jiliang Xu, Xiangjiang Zhan. The establishment of terrestrial vertebrate genetic resource bank and species identification based on DNA barcoding in Wanglang National Nature Reserve [J]. Biodiv Sci, 2023, 31(7): 22661-. |
| [10] | Shengwen Chen, Haibao Ren, Guangrong Tong, Ningning Wang, Wenchao Lan, Jianhua Xue, Xiangcheng Mi. Spatial patterns in woody species diversity in the Qianjiangyuan National Park [J]. Biodiv Sci, 2023, 31(7): 22587-. |
| [11] | Yanqiu Xie, Hui Huang, Chunxiao Wang, Yaqin He, Yixuan Jiang, Zilin Liu, Chuanyuan Deng, Yushan Zheng. Determinants of species-area relationship and species richness of coastal endemic plants in the Fujian islands [J]. Biodiv Sci, 2023, 31(5): 22345-. |
| [12] | Xiuying Zhang, Subinur Eli, Houhun Li. Annual report of new taxa for Chinese Lepidoptera in 2022 [J]. Biodiv Sci, 2023, 31(10): 23283-. |
| [13] | Abdukirim Gulzar, Xiuying Zhang, Subinur Eli, Houhun Li. Annual report of new taxa for Chinese Lepidoptera in 2021 [J]. Biodiv Sci, 2022, 30(8): 22191-. |
| [14] | Ke Yang, Chengzhi Ding, Xiaoyong Chen, Liuyong Ding, Minrui Huang, Jinnan Chen, Juan Tao. Fish diversity and spatial distribution pattern in the Nujiang River Basin [J]. Biodiv Sci, 2022, 30(7): 21334-. |
| [15] | Lujia Tian, Xiaobo Yang, Donghai Li, Long Li, Lin Chen, Caiqun Liang, Peichun Zhang, Chendi Li. Species diversity and nestedness of bird assemblages in the forest fragments of Haikou and Sanya cities [J]. Biodiv Sci, 2022, 30(6): 21424-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2026 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn