



生物多样性 ›› 2019, Vol. 27 ›› Issue (3): 314-326. DOI: 10.17520/biods.2018339 cstr: 32101.14.biods.2018339
杨思琪1,张琪1,宋希强1,王健1,李意德2,许涵2,郭守玉3,丁琼1,*( )
)
收稿日期:2018-12-25
接受日期:2019-02-27
出版日期:2019-03-20
发布日期:2019-04-09
通讯作者:
丁琼
基金资助:
Yang Siqi1,Zhang Qi1,Song Xiqiang1,Wang Jian1,Li Yide2,Xu Han2,Guo Shouyu3,Ding Qiong1,*( )
)
Received:2018-12-25
Accepted:2019-02-27
Online:2019-03-20
Published:2019-04-09
Contact:
Ding Qiong
摘要:
不同功能群的根部真菌可能会与植物差异性地互作, 并进一步影响地下真菌与植物群落构建。本研究采用Illumina Miseq测序方法检测了海南尖峰岭热带山地雨林中常见植物的根部真菌; 采用网络分析法比较了丛枝菌根(AM)真菌、外生菌根(ECM)真菌, 以及所有根部真菌与植物互作的二分网络(bipartite networks)结构特性。从槭树科、番荔枝科、夹竹桃科、冬青科、棕榈科、壳斗科、樟科和木犀科等8科植物的根系中, 检测到297,831条真菌ITS1序列, 这些序列被划为1,279个真菌分类单元(OTUs), 其中子囊菌门748个、担子菌门354个、球囊菌亚门80个, 以及未知真菌97个。核心根部真菌群落(420个OTUs)中, 至少有三类不同生态功能的真菌常见, 即丛枝菌根真菌(40个OTUs, 占总序列数23.4%)、外生菌根真菌(48个OTUs, 13.9%)和腐生型真菌(83个OTUs, 19.8%)。尖峰岭山地雨林根部真菌-植物互作网络结构特性的指标普遍显著高于/低于假定物种随机互作的零模型期待值。在群落水平, 不同功能型的根部真菌-植物互作网络表现出不同或相反的结构特性, 如丛枝菌根互作网络表现为比零模型预测值高的嵌套性和连接性, 以及比零模型低的专一性, 而外生菌根互作网络呈现出比零模型预测值低的嵌套性和连接性, 以及比零模型高的专一性。在功能群水平, 植物的生态位重叠度在AM互作网络高, 而ECM互作网络低; 真菌的生态位宽度在ECM互作网络窄, 而在AM互作网络较宽。共现(co-occurrence)网络分析进一步揭示, ECM群落的物种对资源的高度种间竞争(植物、真菌高C-score), 以及AM群落的物种无明显种间竞争(低C-score), 可能分别是形成反嵌套ECM互作网络及高嵌套AM互作网络结构的原因。上述结果说明, 尖峰岭山地雨林中至少有两种及以上的种间互作机制调节群落构建: 驱动AM互作网络冗余(nestedness)及ECM互作网络的高生态位分化(专一性)。本研究在同一个森林内探讨了不同功能型的真菌-植物互作特性, 对深入理解热带森林的物种共存机制和生态恢复具有重要意义。
杨思琪,张琪,宋希强,王健,李意德,许涵,郭守玉,丁琼 (2019) 尖峰岭热带山地雨林根部真菌-植物互作网络结构特征. 生物多样性, 27, 314-326. DOI: 10.17520/biods.2018339.
Yang Siqi,Zhang Qi,Song Xiqiang,Wang Jian,Li Yide,Xu Han,Guo Shouyu,Ding Qiong (2019) Structural features of root-associated fungus-plant interaction networks in the tropical montane rain forest of Jianfengling, China. Biodiversity Science, 27, 314-326. DOI: 10.17520/biods.2018339.
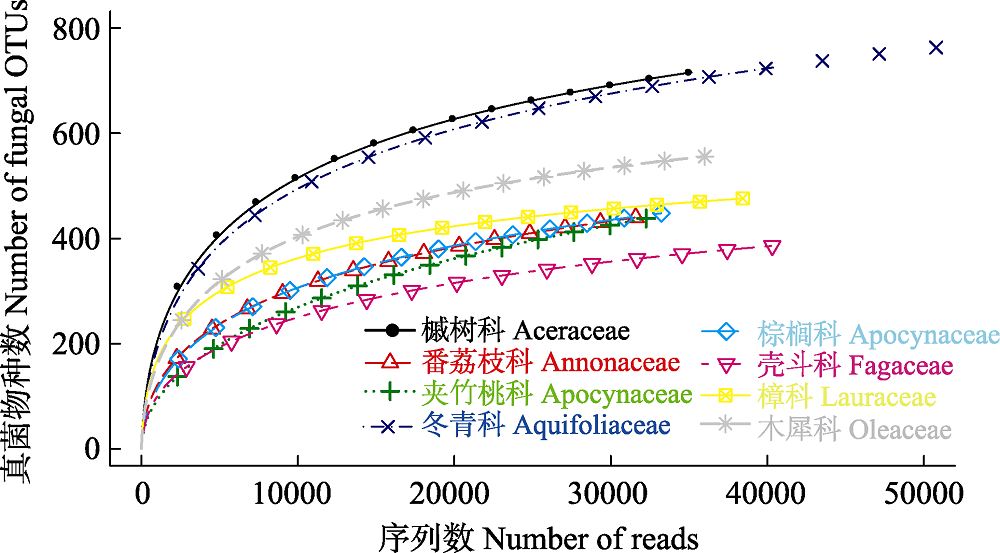
图1 尖峰岭热带山地雨林8科植物根部真菌物种累积曲线
Fig. 1 Rarefaction curves of root-associated fungi of eight plant families in the tropical montane rain forest of Jianfengling
| 宿主植物 Host plant | 根样品数 No. of root samples | 真菌物种数 / reads No. of fungal species/reads | 核心真菌物种数 / reads No. of core fungal species/reads | Shannon多样性指数 Shannon diversity index |
|---|---|---|---|---|
| 槭树科 Aceraceae | 34 | 701 / 31,589 | 328 / 29,817 | 3.97 |
| 番荔枝科 Annonaceae | 32 | 440 / 31,588 | 256 / 30,846 | 2.88 |
| 夹竹桃科 Apocynaceae | 41 | 434 / 31,589 | 277 / 31,134 | 2.58 |
| 冬青科 Aquifoliaceae | 39 | 675 / 31,588 | 334 / 29,932 | 4.05 |
| 棕榈科 Arecaceae | 21 | 436 / 31,589 | 264 / 30,894 | 3.12 |
| 壳斗科 Fagaceae | 31 | 359 / 31,589 | 217 / 30,983 | 2.58 |
| 樟科 Lauraceae | 35 | 463 / 31,589 | 261 / 30,508 | 4.04 |
| 木犀科 Oleaceae | 34 | 546 / 31,589 | 302 / 30,527 | 3.72 |
表1 尖峰岭热带山地雨林8科植物的根部真菌群落物种丰富度和多样性
Table 1 Species diversity and richness of root-associated fungal community of eight plant families in the tropical montane rain forest of Jianfengling
| 宿主植物 Host plant | 根样品数 No. of root samples | 真菌物种数 / reads No. of fungal species/reads | 核心真菌物种数 / reads No. of core fungal species/reads | Shannon多样性指数 Shannon diversity index |
|---|---|---|---|---|
| 槭树科 Aceraceae | 34 | 701 / 31,589 | 328 / 29,817 | 3.97 |
| 番荔枝科 Annonaceae | 32 | 440 / 31,588 | 256 / 30,846 | 2.88 |
| 夹竹桃科 Apocynaceae | 41 | 434 / 31,589 | 277 / 31,134 | 2.58 |
| 冬青科 Aquifoliaceae | 39 | 675 / 31,588 | 334 / 29,932 | 4.05 |
| 棕榈科 Arecaceae | 21 | 436 / 31,589 | 264 / 30,894 | 3.12 |
| 壳斗科 Fagaceae | 31 | 359 / 31,589 | 217 / 30,983 | 2.58 |
| 樟科 Lauraceae | 35 | 463 / 31,589 | 261 / 30,508 | 4.04 |
| 木犀科 Oleaceae | 34 | 546 / 31,589 | 302 / 30,527 | 3.72 |
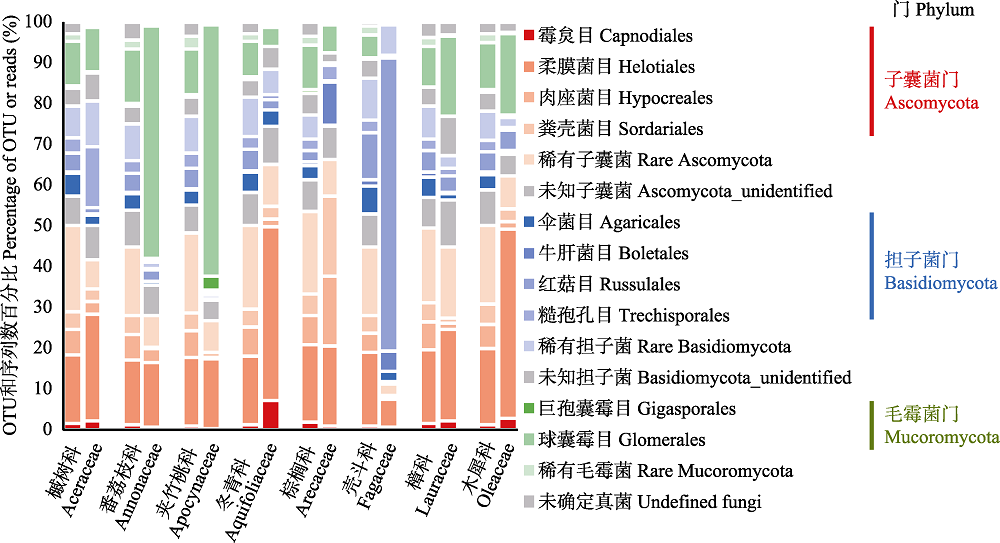
图2 尖峰岭热带山地雨林8科植物的根部真菌目组成差异。每科植物对应的左柱与右柱分别为OTUs和reads百分比。
Fig. 2 Compositional differences of root-associated fungi by order among the eight plant families in the tropical montane rain forest of Jianfengling. For each plant family, left and right column are percentage of OTUs and reads, respectively.
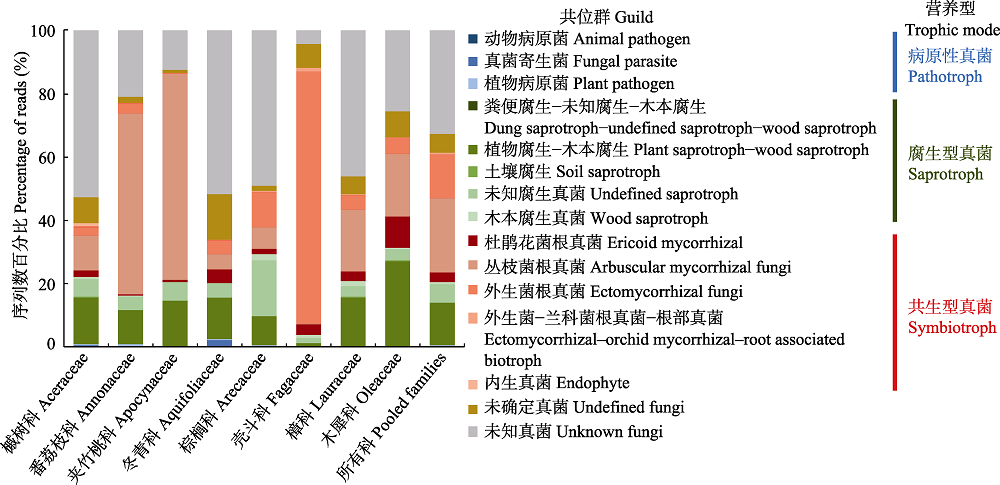
图3 尖峰岭热带山地雨林8科植物的根部不同共位群和营养型真菌的序列百分比组成差异
Fig. 3 Compositional differences in reads of root-associated fungi by guilds and trophic modes of the eight plant families in the tropical montane rain forest of Jianfengling
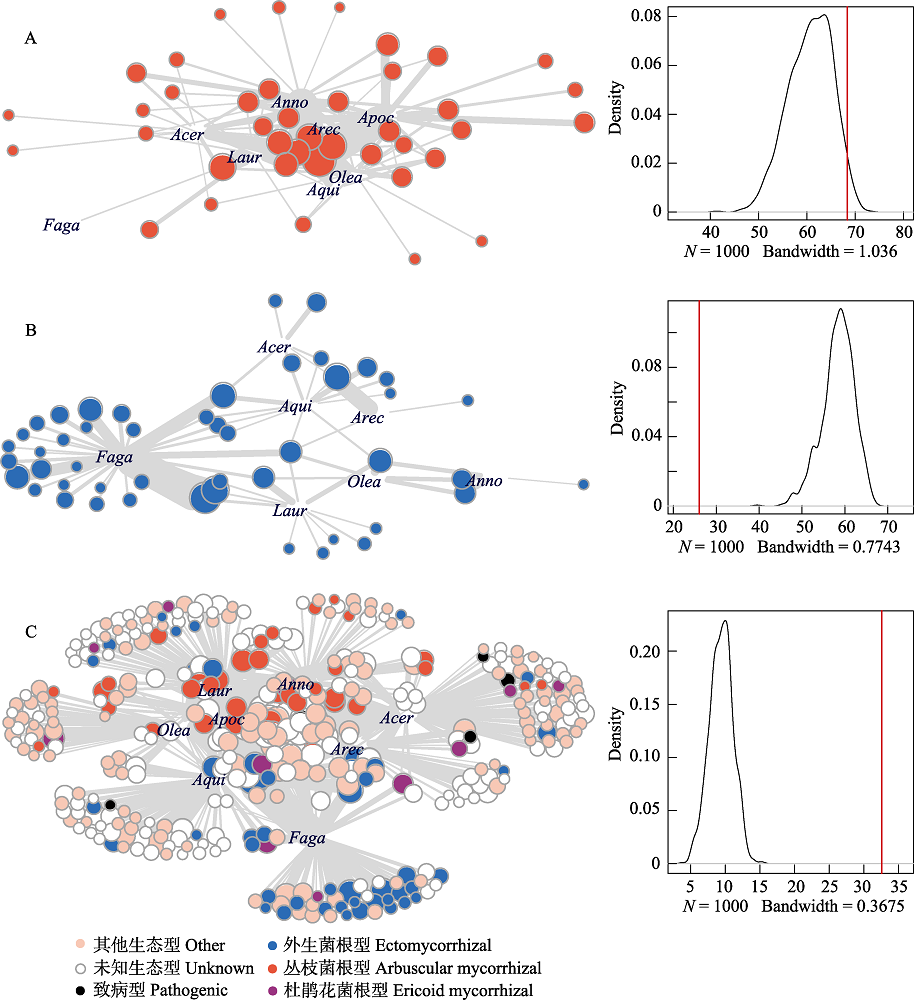
图4 尖峰岭热带山地雨林根部真菌与植物的种间相互作用网络结构(左)与嵌套性(NODF)密度函数图及嵌套性实际观察值 (红色垂直线)(右)。(A)丛枝菌根互作网络; (B)外生菌根互作网络; (C)所有根部真菌-植物互作网络。Acer: 槭树科; Anno: 番荔枝科; Apoc: 夹竹桃科; Aqui: 冬青科; Arec: 棕榈科; Faga: 壳斗科; 灰色连线代表真菌与宿主的种间互作, 线宽对应互作强弱。
Fig. 4 Root-associated fungus-plant interaction networks (left) with density plots showing the distribution of nestedness (NODF) predicted by Patefiled’s null model (right) and observed nestedness (red vertical line) in tropical montane rain forest of Jianfenling Mountain Hainan Island. (A) and (B) are partial networks of AM and ECM interactions, and (C) is the whole root-associated fungus-plant interaction network. Acer, Aceraceae; Anno, Annoaceae; Apoc, Apocynaceae; Aqui, Aquifoliaceae; Arec, Arecaceae; Faga, Fagaceae; Laur, Lauraceae; Olea, Oleaceae. Interactions between plant and fungi are indicated by grey lines with thickness proportional to interaction strength.
| 丛枝菌根网络 Arbuscular mycorrhizal network | 外生菌根网络 Ectomycorrhizal network | 根部真菌-植物互作网络 Root-associated fungus-plant network | ||||
|---|---|---|---|---|---|---|
| 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | |
| 网络嵌套性 Nestedness metric based on overlap and decreasing fill, NODF | 68.39 | 60.70 | 26.06 | 58.36 | 32.59 | 9.40 |
| 网络加权嵌套性 Weighted NODF | 45.12 | 53.76 | 18.04 | 48.44 | 20.46 | 7.69 |
| 网络专一性 Specialization (H°2) | 0.31 | 0.01 | 0.81 | 0.01 | 0.54 | 0.01 |
| 网络连接性 Connectance | 0.42 | 0.88 | 0.21 | 0.89 | 0.26 | 0.99 |
| 植物生态位重叠 Niche overlap of plants | 0.46 | 0.99 | 0.11 | 0.99 | 0.25 | 0.99 |
| 真菌生态位重叠 Niche overlap of fungi | 0.35 | 0.97 | 0.39 | 0.98 | 0.19 | 0.91 |
| 植物的伙伴多样性 Generality of plants | 5.46 | 7.87 | 5.32 | 10.92 | 34.14 | 87.31 |
| 真菌的伙伴多样性 Vulnerability of fungi | 3.41 | 4.84 | 1.28 | 2.74 | 3.16 | 7.95 |
| 植物Checkboard值 C-score of plants | 0.17 | 0.28 | 0.62 | 0.48 | 0.38 | 0.96 |
| 真菌Checkboard值 C-score of fungi | 0.24 | 0.02 | 0.52 | 0.43 | 0.62 | 0.85 |
表2 根部真菌-植物互作网络结构特性指数
Table 2 Structural features of root-associated fungus-plant interaction networks
| 丛枝菌根网络 Arbuscular mycorrhizal network | 外生菌根网络 Ectomycorrhizal network | 根部真菌-植物互作网络 Root-associated fungus-plant network | ||||
|---|---|---|---|---|---|---|
| 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | 观察值 Observed | 零模型 Null model | |
| 网络嵌套性 Nestedness metric based on overlap and decreasing fill, NODF | 68.39 | 60.70 | 26.06 | 58.36 | 32.59 | 9.40 |
| 网络加权嵌套性 Weighted NODF | 45.12 | 53.76 | 18.04 | 48.44 | 20.46 | 7.69 |
| 网络专一性 Specialization (H°2) | 0.31 | 0.01 | 0.81 | 0.01 | 0.54 | 0.01 |
| 网络连接性 Connectance | 0.42 | 0.88 | 0.21 | 0.89 | 0.26 | 0.99 |
| 植物生态位重叠 Niche overlap of plants | 0.46 | 0.99 | 0.11 | 0.99 | 0.25 | 0.99 |
| 真菌生态位重叠 Niche overlap of fungi | 0.35 | 0.97 | 0.39 | 0.98 | 0.19 | 0.91 |
| 植物的伙伴多样性 Generality of plants | 5.46 | 7.87 | 5.32 | 10.92 | 34.14 | 87.31 |
| 真菌的伙伴多样性 Vulnerability of fungi | 3.41 | 4.84 | 1.28 | 2.74 | 3.16 | 7.95 |
| 植物Checkboard值 C-score of plants | 0.17 | 0.28 | 0.62 | 0.48 | 0.38 | 0.96 |
| 真菌Checkboard值 C-score of fungi | 0.24 | 0.02 | 0.52 | 0.43 | 0.62 | 0.85 |
| 1 |
Almario J, Jeena G, Wunder J, Langen G, Zuccaro A, Coupland G, Bucher M ( 2017) Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proceedings of the National Academy of Sciences, USA, 114, 9403-9412.
DOI URL PMID |
| 2 |
Almeida-Neto M, Ulrich W ( 2011) A straightforward computational approach for measuring nestedness using quantitative matrices. Environmental Modelling and Software, 26, 173-178.
DOI URL |
| 3 |
Bahram M, Peay KG, Tedersoo L ( 2014) Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytologist, 205, 1454-1463.
DOI URL PMID |
| 4 |
Bascompte J ( 2007) Networks in ecology. Basic and Applied Ecology, 8, 485-490.
DOI URL |
| 5 |
Bascompte J, Jordano P, Melián CJ, Olesen JM ( 2003) The nested assembly of plant-animal mutualistic networks. Proceedings of the National Academy of Sciences, USA, 100, 9383-9387.
DOI URL PMID |
| 6 |
Bastolla U, Fortuna MA, Pascual-Garcia A, Ferrera A, Luque B, Bascompte J ( 2009) The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature, 458, 1018-1020.
DOI URL |
| 7 |
Blaalid R, Davey ML, Kauserud H, Carlsen T, Halvorsen R, Hoiland K, Eidesen PB ( 2014) Arctic root-associated fungal community composition reflects environmental filtering. Molecular Ecology, 23, 649-659.
DOI URL PMID |
| 8 |
Blüthgen N, Fründ J, Vázquez DP, Menzel F ( 2008) What do interaction network metrics tell us about specialization and biological traits. Ecology, 89, 3387-3399.
DOI URL PMID |
| 9 |
Burkle LA, Marlin JC, Knight TM ( 2013) Plant-pollinator interactions over 120 years: Loss of species, co-occurrence, and function. Science, 339, 1611-1615.
DOI URL PMID |
| 10 | Cannon PF, Kirk PM ( 2007) Fungal Families of the World. CABI Bioscience, Wallingford. |
| 11 |
Cardoso EJBN, Nogueira MA, Zangaro W ( 2017) Importance of mycorrhizae in tropical soils. In: Diversity and Benefits of Microorganisms from the Tropics (eds de Azevedo J, Quecine M), Springer, Cham.
DOI URL |
| 12 |
Chagnon PL, Bradley RL, Klironomos JN ( 2012) Using ecological network theory to evaluate the causes and consequences of arbuscular mycorrhizal community structure. New Phytologist, 194, 307-312.
DOI URL |
| 13 |
Chen L, Zheng Y, Gao C, Mi XC, Ma KP, Wubet T, Guo LD ( 2017) Phylogenetic relatedness explains highly interconnected and nested symbiotic networks of woody plants and arbuscular mycorrhizal fungi in a Chinese subtropical forest. Molecular Ecology, 26, 2563-2575.
DOI URL PMID |
| 14 |
Corrales A, Henkel TW, Smith ME ( 2018) Ectomycorrhizal associations in the tropics—Biogeography, diversity patterns and ecosystem roles. New Phytologist, 220, 1076-1091.
DOI URL PMID |
| 15 |
Dickie KH, Cann C, Norman EC, Bamforth CW, Muller RE ( 2001) Foam-negative materials. Journal of the American Society of Brewing Chemists, 59, 17-23.
DOI URL |
| 16 |
Edgar RC ( 2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460-2461.
DOI URL |
| 17 |
Edgar RC ( 2013) UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10, 996.
DOI URL PMID |
| 18 |
Edgar RC ( 2018) Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences, PeerJ, 6, e4652.
DOI URL |
| 19 |
Fang JY, Li YD, Zhu B, Liu GH, Zhou GY ( 2004) Community structures and species richness in the montane rain forest of Jianfengling, Hainan Island, China. Biodiversity Science, 12, 29-43. (in Chinese with English abstract)
DOI URL |
|
[ 方精云, 李意德, 朱彪, 刘国华, 周光益 ( 2004) 海南岛尖峰岭山地雨林的群落结构、物种多样性以及在世界雨林中的地位. 生物多样性, 12, 29-43.]
DOI URL |
|
| 20 |
Fortuna MA, Stouffer DB, Olesen JM, Jordano P, Mouillot D, Krasnov BR, Poulin R, Bascompte J ( 2010) Nestedness versus modularity in ecological networks: Two sides of the same coin? Journal of Animal Ecology, 79, 811-817.
DOI URL PMID |
| 21 |
Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Hutmacher RB, Dahlberg JA, Coleman-Derr D, Lemaux PG, Taylor JW ( 2019) Strong succession in arbuscular mycorrhizal fungal communities. The ISME Journal, 13, 214-226.
DOI |
| 22 |
Gotelli NJ, McCabe DJ ( 2002) Species co-occurrence: A meta-analysis of J. M. Diamond’s assembly rules model. Ecology, 83, 2091-2096.
DOI URL |
| 23 |
Huang CW, Liao YH, Ding Q ( 2017) Two sample pooling strategies revealed different root-associated fungal diversity of Rhododendron species. Acta Microbiologica Sinica, 57, 571-581. (in Chinese with English abstract)
DOI URL |
|
[ 黄彩微, 廖映辉, 丁琼 ( 2017) 两种混合样品策略对揭示杜鹃花根部真菌多样性的影响. 微生物学报, 57, 571-581.]
DOI URL |
|
| 24 |
James A, Pitchford JW, Plank MJ ( 2012) Disentangling nestedness from models of ecological complexity. Nature, 487, 227-230.
DOI URL PMID |
| 25 | Jiang YX, Lu JP ( 1991) Tropical Forest Ecosystem of Jianfengling, Hainan Island, China. Science Press, Beijing. (in Chinese) |
| [ 蒋有绪, 卢俊培 ( 1991) 中国海南岛尖峰岭热带林生态系统. 科学出版社, 北京.] | |
| 26 |
Kivlin SN, Winston GC, Goulden ML, Treseder KK ( 2014) Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecology, 12, 14-25.
DOI URL |
| 27 |
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüβler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H ( 2013) Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology, 22, 5271-5277.
DOI URL PMID |
| 28 |
Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E ( 2009) Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences, USA, 106, 18621-18626.
DOI URL |
| 29 |
Merges D, Bálint M, Schmitt I, Böhning-Gaese K, Neuschulz EL ( 2018) Spatial patterns of pathogenic and mutualistic fungi across the elevational range of a host plant. Journal of Ecology, 106, 1545-1557.
DOI URL |
| 30 |
Newsham KK ( 2011) A meta-analysis of plant responses to dark septate root endophytes. New Phytologist, 190, 783-793.
DOI URL |
| 31 |
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG ( 2016) FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology, 20, 241-248.
DOI URL |
| 32 |
Patefield WM ( 1981) Algorithm AS 159: An efficient method of generating random R × C tables with given row and column totals. Journal of the Royal Statistical Society, Series C (Applied Statistics), 30, 91-97.
DOI URL |
| 33 |
Pianka ER ( 1974) Niche overlap and diffuse competition. Proceedings of the National Academy of Sciences, USA, 71, 2141-2145.
DOI URL PMID |
| 34 | Smith DP, Peay KG ( 2014) Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE, 9, e90234. |
| 35 | Smith SE, Read DJ ( 2008) Mycorrhizal Symbiosis, 3rd edn. Academic Press, London. |
| 36 |
Taudiere A, Munoz F, Lesne A, Monnet AC, Bellanger JM, Selosse MA, Moreau PA, Richard F ( 2015) Beyond ectomycorrhizal bipartite networks: Projected networks demonstrate contrasted patterns between early- and late-successional plants in Corsica. Frontiers in Plant Science, 6, 881.
DOI URL PMID |
| 37 | Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, Kesel AD, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K ( 2014) Global diversity and geography of soil fungi. Science, 346, 1078-1088. |
| 38 |
Thebault E, Fontaine C ( 2010) Stability of ecological communities and the architecture of mutualistic and trophic Networks. Science, 329, 853-856.
DOI URL PMID |
| 39 |
Thiéry O, Vasar M, Jairus T, Davison J, Roux C, Kivistik PA, Metspalu A, Milani L, Saks ü, Moora M, Zobel M, Öpik M ( 2016) Sequence variation in nuclear ribosomal small subunit, internal transcribed spacer and large subunit regions of Rhizophagus irregularis and Gigaspora margarita is high and isolate-dependent. Molecular Ecology, 25, 2816-2832.
DOI URL |
| 40 |
Toju H, Sato H, Yamamoto S, Kadowaki K, Tanabe AS, Yazawa S, Nishimura O, Agata K ( 2013 a) How are plant and fungal communities linked to each other in belowground ecosystems? A massively parallel pyrosequencing analysis of the association specificity of root-associated fungi and their host plants. Ecology and Evolution, 3, 3112-3124.
DOI URL PMID |
| 41 |
Toju H, Yamamoto S, Sato H, Tanabe AS, Gilbert GS, Kadowaki K ( 2013 b) Community composition of root-associated fungi in a Quercus-dominated temperate forest: “Codominance” of mycorrhizal and root-endophytic fungi. Ecology and Evolution, 3, 1281-1293.
DOI URL |
| 42 |
Tylianakis JM ( 2009) Warming up food webs. Science, 323, 1300-1301.
DOI URL |
| 43 |
Ulrich W, Gotelli NJ ( 2007) Null model analysis of species nestedness patterns. Ecology, 88, 1824-1831.
DOI URL PMID |
| 44 |
Vázquez DP, Chacoff NP, Cagnolo L ( 2009) Evaluating multiple determinants of the structure of plant-animal mutualistic networks. Ecology, 90, 2039-2046.
DOI URL PMID |
| 45 |
Xu H, Li YD, Lin MX, Wu JH, Luo TS, Zhou Z, Chen DX, Yang H, Li GJ, Liu SR ( 2015) Community characteristics of a 60 ha dynamics plot in the tropical montane rain forest in Jianfengling, Hainan Island. Biodiversity Science, 23, 192-201. (in Chinese with English abstract)
DOI URL |
|
[ 许涵, 李意德, 林明献, 吴建辉, 骆土寿, 周璋, 陈德祥, 杨怀, 李广建, 刘世荣 ( 2015) 海南尖峰岭热带山地雨林60 ha动态监测样地群落结构特征. 生物多样性, 23, 192-201.]
DOI URL |
|
| 46 |
Yamamoto S, Sato H, Tanabe AS, Hidaka A, Kadowaki K, Toju H ( 2014) Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS ONE, 9, e96363.
DOI URL PMID |
| 47 |
Zhang J, Kobert K, Flouri T, Stamatakis A ( 2013) PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics, 30, 614-620.
DOI URL PMID |
| [1] | 胡志清, 董路. 城市化对鸟类参与的种间互作的影响[J]. 生物多样性, 2024, 32(8): 24048-. |
| [2] | 冯志荣, 陈明波, 杨小芳, 王刚, 董乙乂, 彭艳琼, 陈华燕, 王波. 榕树繁育系统及榕小蜂资源利用塑造了榕小蜂群落[J]. 生物多样性, 2024, 32(3): 23307-. |
| [3] | 王朝雅, 李金涛, 刘畅, 王波, 苗白鸽, 彭艳琼. 西双版纳热带植物园蝴蝶多样性稳定的年际变化及幼虫与植物的互作网络结构[J]. 生物多样性, 2023, 31(12): 23305-. |
| [4] | 李国华, 郭向阳, 李霖明, 任明迅, 万玲, 丁琼, 李娟玲. 海南热带雨林国家公园不同植被类型的大型真菌多样性[J]. 生物多样性, 2022, 30(7): 22110-. |
| [5] | 王剑, 董乙乂, 马丽滨, 潘勃, 马方舟, 丁晖, 胡亚萍, 彭艳琼, 吴孝兵, 王波. 西双版纳国家级自然保护区蚂蚁-树互作网络空间变异[J]. 生物多样性, 2020, 28(6): 695-706. |
| [6] | 许格希, 史作民, 唐敬超, 许涵, 杨怀, 刘世荣, 李意德, 林明献. 物种多度和径级尺度对于评价群落系统发育结构的影响: 以尖峰岭热带山地雨林为例[J]. 生物多样性, 2016, 24(6): 617-628. |
| [7] | 陈艳, 李宏庆, 刘敏, 陈小勇. 榕-传粉榕小蜂间的专一性与协同进化[J]. 生物多样性, 2010, 18(1): 1-10. |
| [8] | 邵士成, 吴少华, 陈有为, 王立东, 杨丽源, 李绍兰, 李治滢. 云南元江印楝植物内生真菌的种类组成[J]. 生物多样性, 2008, 16(1): 63-67. |
| [9] | 欧芷阳, 杨小波, 吴庆书. 尖峰岭自然保护区扩大区域植物多样性研究[J]. 生物多样性, 2007, 15(4): 437-444. |
| [10] | 王文进, 张明, 刘福德, 郑建伟, 王中生, 张世挺, 杨文杰, 安树青. 海南岛吊罗山热带山地雨林两个演替阶段的种间联结性[J]. 生物多样性, 2007, 15(3): 257-263. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn