



生物多样性 ›› 2011, Vol. 19 ›› Issue (1): 3-16. DOI: 10.3724/SP.J.1003.2011.14256 cstr: 32101.14.SP.J.1003.2011.14256
所属专题: 昆虫多样性与生态功能
收稿日期:2010-10-26
接受日期:2010-12-30
出版日期:2011-01-20
发布日期:2011-04-01
通讯作者:
孔宏智
作者简介:*E-mail: hzkong@ibcas.ac.cn基金资助:
An Li1,2, Guixia Xu1, Hongzhi Kong1,*( )
)
Received:2010-10-26
Accepted:2010-12-30
Online:2011-01-20
Published:2011-04-01
Contact:
Hongzhi Kong
摘要:
拷贝数目变异是一种对表型变异和生物进化具有重要意义的基因组结构变异。以前的研究表明不同物种中F-box基因的拷贝数目差异较大。为了深入探索拷贝数目变异的式样和机制, 我们以12个果蝇近缘种为研究对象, 分析了F-box基因的系统发育关系、进化式样以及它们在染色体上的位置。结果发现, 虽然各个物种中F-box基因的拷贝数目差别不大(42-47个), 但是仍然存在着很多引起拷贝数目变异的基因获得和丢失事件。这说明表面上变化不大的拷贝数目在一定程度上掩盖了频繁发生的基因获得和丢失事件。通过比较这些基因在染色体上的位置, 发现只有在亲缘关系很近的物种之间才能鉴定出有明显微共线性关系的基因组区段。我们还发现, 造成F-box基因拷贝数目增加的主要机制是散在重复和串联重复, 而反转录转座和新基因的非编码区起源也是两种值得注意的机制。此外, 序列变异导致的外显子边界变化以及外显子丢失是引起拷贝数目减少的两种机制。在12种果蝇的最近共同祖先中, F-box基因的拷贝数目与现存物种基本相似, 但是基因的获得和丢失事件使得现存物种中的F-box基因在构成上已经有了明显的差别。对数目变异的式样及其与基因功能的关系的研究表明, 拷贝数目变异是F-box基因家族“生与死”的进化在基因组层面的系统反映, 并有可能为表型变异提供了原始材料。
李安, 徐桂霞, 孔宏智 (2011) F-box基因拷贝数目变异的机制研究:以12种果蝇为例. 生物多样性, 19, 3-16. DOI: 10.3724/SP.J.1003.2011.14256.
An Li, Guixia Xu, Hongzhi Kong (2011) Mechanisms underlying copy number variation in F-box genes: evidence from comparison of 12 Drosophila species. Biodiversity Science, 19, 3-16. DOI: 10.3724/SP.J.1003.2011.14256.
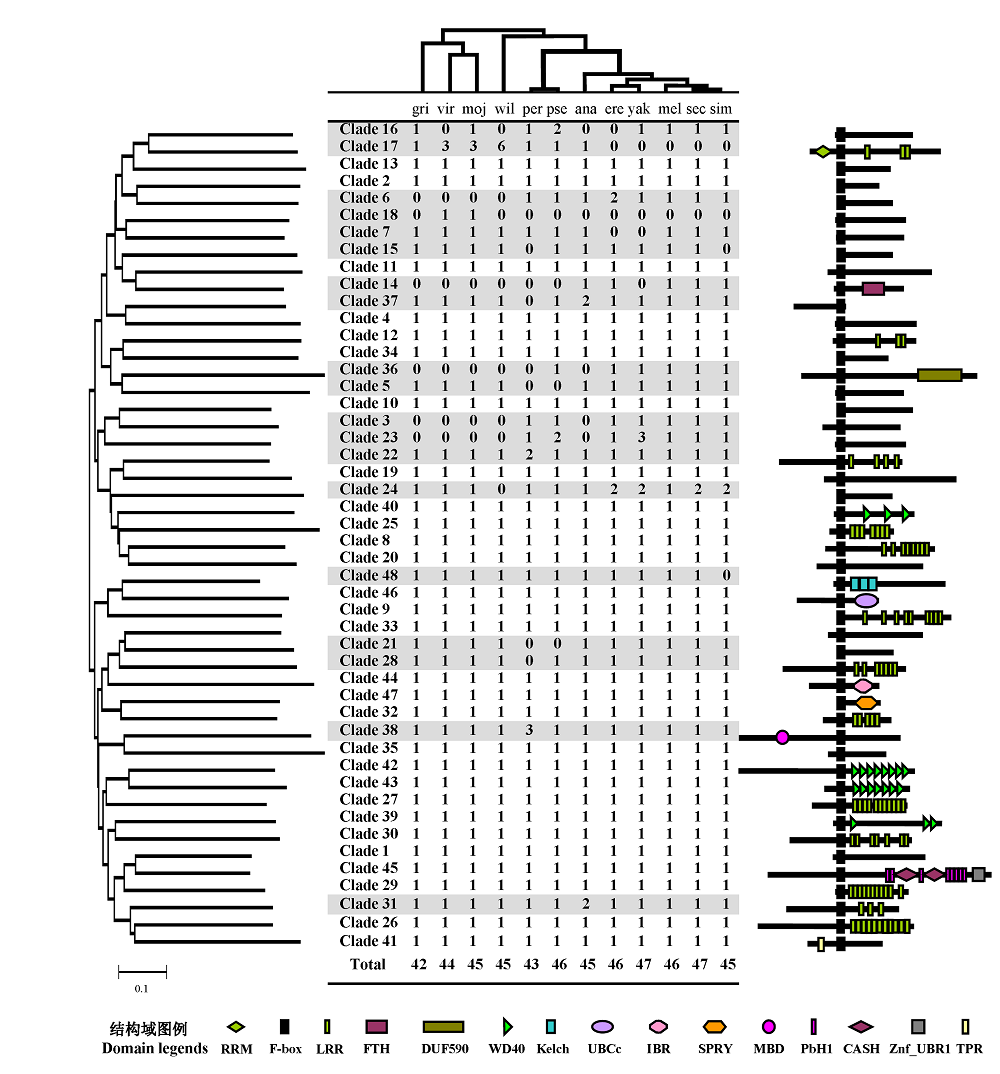
图1 果蝇12个近缘种的F-box基因的拷贝数目和对应的结构域组成。统计表的上方是果蝇12个近缘种的物种树(修改自 http://rana.lbl.gov/drosophila/index.html), 分支末端对应各自的物种名(用种名的前三个字母代替), 即拷贝数目统计表的第一行。统计表左侧是用48个直系同源基因分支中的代表序列构建的系统树, 各支末端对应统计表中该基因在12个物种中的拷贝数目; 右侧则为该基因编码的蛋白质的结构域组成模式图。灰色标出的行是有拷贝数目变异的基因。
Fig. 1 Copy number and domain structure of F-box proteins from 12 Drosophila species. The phylogenetic tree of the 12 Drosophila species (modified from http://rana.lbl.gov/drosophila/index.html) is above the table. The first three letters of each specific epithet are used as the abbreviation for each species. On the left side of the table is a neighbor-joining (NJ) tree for 48 clades of F-box proteins whose domain structure are shown on the right. Numbers in the table indicate the copy numbers of F-box genes belonging to each clade. Clades with copy number variations are shaded.
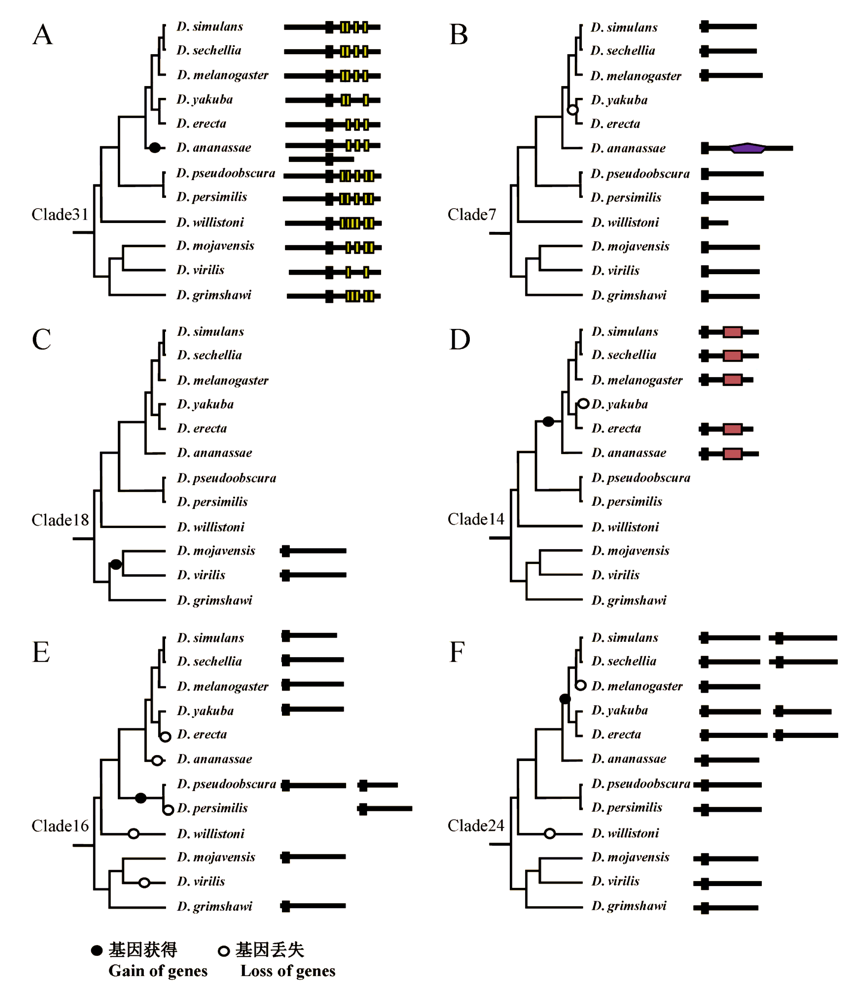
图2 F-box基因拷贝数目变异的典型情况。结构域的图例与图1的一致。A: 该基因在12个物种中均存在, 且在D. ananassae中又获得了一个拷贝; B: 该基因存在于10个物种中, D. yakuba和D. erecta的最近共同祖先丢失了这个基因; C: 该基因仅在D. mojavensis和D. virilis中存在, 说明此基因是在这两个物种的最近共同祖先中获得的; D: 物种树上部6个物种的最近共同祖先获得了该基因, 又在D. yakuba中发生丢失。E: 该基因在D. erecta、D. ananassae、D. willistoni和D. virilis这4个物种中分别丢失, 而在D. pseudoobscura和D. persimilis的最近共同祖先中发生了基因重复, 产生了第二个拷贝, 后来D. persimilis中的第一个拷贝丢失; F: 该基因在11个物种中存在, 在D. willistoni中丢失。物种树上部5个物种的最近共同祖先中增加了1个拷贝, 但D. melanogaster又丢掉了1个拷贝。
Fig. 2 Examples of copy number variation of F-box genes. Domain legends are the same as those in A, D. ananassae gained a new copy of F-box gene; B, The most recent common ancestor (MRCA) of D.yakuba and D.erecta lost the ortholog of Clade7; C, MRCA of D.mojavensisandD.virilis gained a new gene; D, MRCA of the top six species gained a new copy of F-box gene and D.yakuba lost this gene after their divergence; E, Four gene loss events occurred independently in D. erecta, D. ananassae, D. willistoni, and D. virilis. A gene duplication event happened in the MRCA of D.pseudoobscura and D.persimilis, generating two copies, but then one copy lost in D.persimilis; F, D.willistoni lost Clade24 gene. The MRCA of the top five species gained a new copy of this gene, but then D.melanogaster lost the new copy.
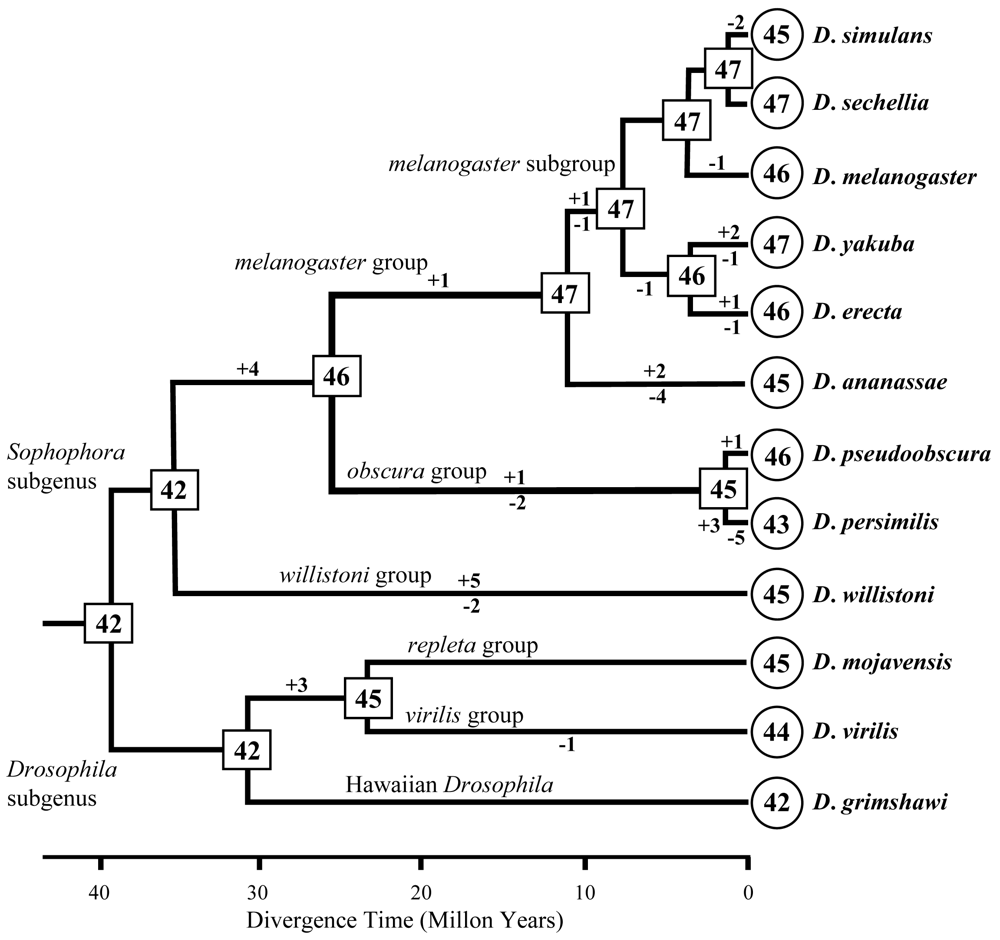
图3 F-box基因的拷贝数目在果蝇进化历史中的变化。圆圈和方框中的数字分别表示12个现存物种及其祖先中的拷贝数目。系统树分支上的+/-及数字表示基因拷贝数目的增减事件及增减的拷贝数目。系统发育树修改自Assembly/Alignment/ Annotation of 12 related Drosophila species [http://rana.lbl.gov/drosophila/index.html]。
Fig. 3 Evolutionary change of the copy number of F-box genes in Drosophila species. The numbers in circles and rectangles represent the copy numbers of genes in extant and ancestral species, respectively. Numbers above and below each branch indicate the numbers of gains (+) and losses (-) of genes, respectively. The phylogenetic tree is modified from Assembly/Alignment/Annotation of 12 related Drosophila species [http://rana.lbl.gov/drosophila/index.html].
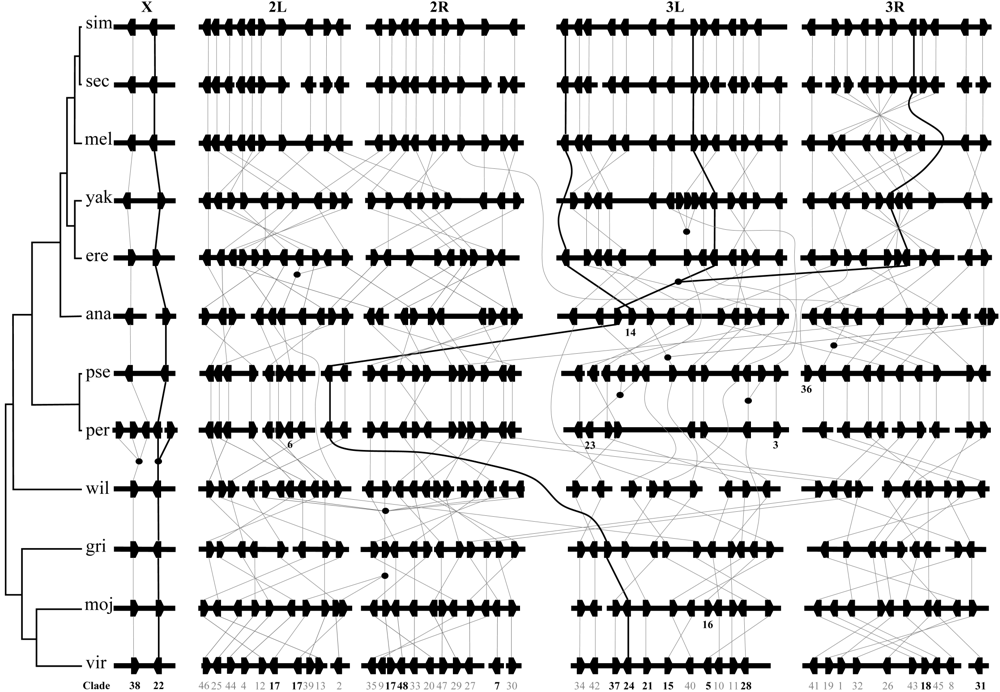
图4 12个果蝇近缘种基因组中F-box基因的染色体定位及相互之间的直系同源与旁系同源关系。横向的长条表示12个果蝇的染色体臂/支架/片段。L和R分别指示染色体的左、右臂。五边形的黑色方块表示F-box基因。直系同源基因之间用直线相连, 实心圆圈表示基因重复事件。曲线表示其所穿过的染色体上已经丢失相应的直系同源基因。基因下面的数字代表其所在进化支的编号(粗体表示有拷贝数目变异的进化支)。由于这12个种的核型不同, 我们在D. melanogaster中将基因按从X染色体到3号染色体右臂的顺序展示, 并将其他物种的染色体按与D. melanogaster相对应的顺序排列。加粗的线指示了拷贝数目变异的三个例子。分别包括了一次获得(Clade22)、一次获得和一次丢失(Clade14)以及一次获得和两次丢失(Clade24)事件。
Fig. 4 Chromosomal locations of F-box genes and their orthologous and paralogous relationships in the 12 Drosophila genomes. Horizontal bars represent the chromosomal arms/scaffolds/segments of the 12 Drosophila genomes. L and R indicate the left and right arms of chromosomes, respectively. Hexagonal lumps designate F-box genes. Orthologs are connected by lines. Filled circles indicate gene duplication events. Orthologs connected by curve do not exist in the species between them. Numbers below genes are the No. of clades (those in bold indicate that the copy numbers are different among the 12 species). Since the 12 species are different in karyotype, we arranged the genes from chromosomes X to 3R in D. melanogaster, and the chromosomes in other species are arranged corresponding to D. melanogaster. Lines in bold indicate three examples of copy number variation, which show one gain (Clade22), one gain and one loss (Clade14), as well as one gain and two losses (Clade24), respectively.
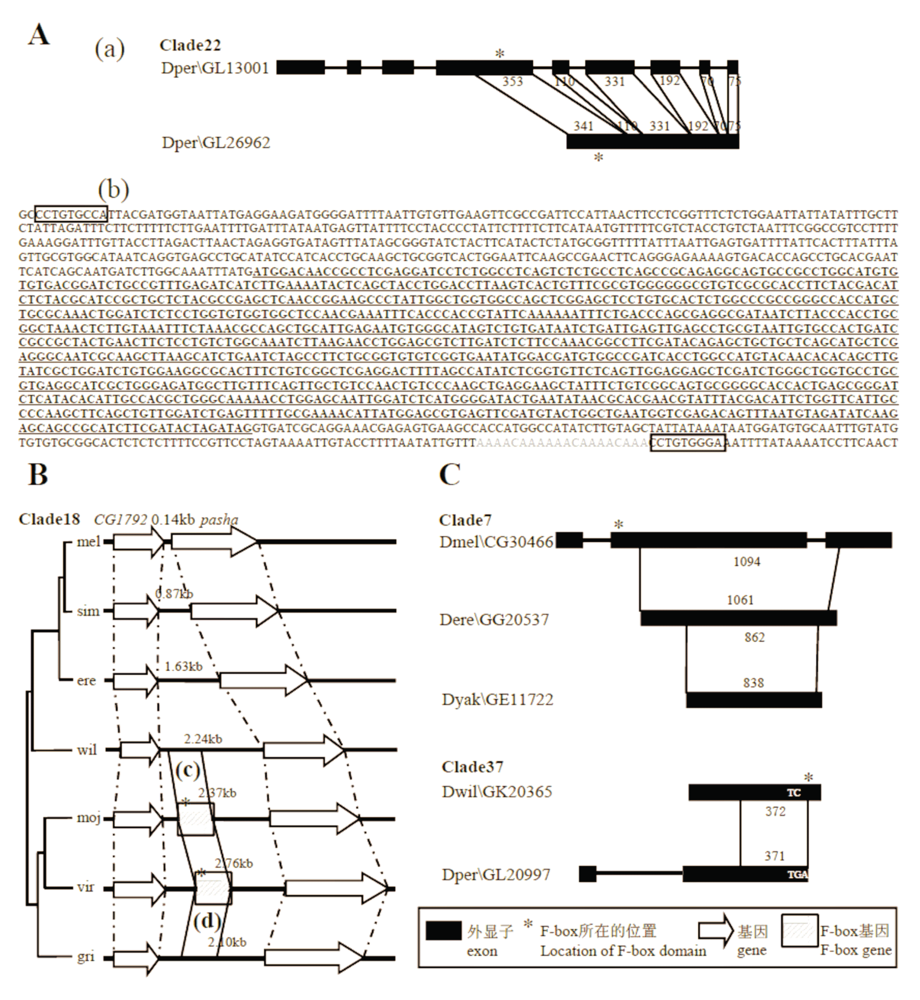
图5 F-box基因拷贝数目变异的机制。A: Clade22拷贝数目增加的机制是反转录转座。(a)旁系同源基因的结构比较(祖先状态在上)。细线连接具有较高序列一致性的部分, 数字表示该部分的长度。(b)该无内含子的F-box基因(下划线且粗体)有一个类似poly-A尾巴(灰色)的结构和两段正向重复序列(框内)。B: Clade18拷贝数目增加的机制是从头起源。新基因起源于CG1792和pasha(D. melanogaster中的名字)两基因之间的非编码序列。(c)和(d)是该F-box基因与前述非编码序列的比对示意图(详见附录V)。C: F-box基因拷贝数目减少的机制。图示直系同源基因之间的结构比较(祖先状态在上)。
Fig. 5 Mechanisms for copy number variation of F-box genes. A, The mechanism that caused the increase in copy number of Clade22 was retroposition. (a) Comparison of the exon/intron structure of paralogs is showed (the upper gene is similar to ancestral gene). Regions that can match to each other are connected with thin lines, and the numbers show the length of the matched parts. (b) The intronless F-box gene (underlined and in bold) likely possesses a poly-A tail (in gray) and two direct repeats (in boxes). B, The mechanism that caused the increase in copy number of Clade18 was de novo origination. The new gene was derived from non-coding sequences between CG1792 and pasha (respectively named after orthologous genes in D. melanogaster). (c) and (d) are alignments between non-coding sequences and F-box genes (Appendix V for the details). C, Mechanisms for the decrease in copy number of F-box genes. Comparison of the exon/intron structures between orthologs are showed (the upper gene is ancestral).
| 冗余序列 Redundant sequences | 所选的序列 Selected sequences |
|---|---|
| FBpp0073103, FBpp0073101, FBpp0073102 | FBpp0073102 |
| FBpp0078592, FBpp0110535, FBpp0078594, FBpp0078593 | FBpp0078593 |
| FBpp0084796, FBpp0084795 | FBpp0084795 |
| FBpp0110196, FBpp0078693 | FBpp0078693 |
| FBpp0087190, FBpp0111982, FBpp0111980, FBpp0111981 | FBpp0111981 |
| FBpp0086875, FBpp0086876 | FBpp0086876 |
| FBpp0087274, FBpp0087275, FBpp0087276 | FBpp0087276 |
| FBpp0083216, FBpp0083215 | FBpp0083215 |
| FBpp0099839, FBpp0099383 | FBpp0099383 |
| FBpp0268227, FBpp0264637 | FBpp0264637 |
| FBpp0259602, FBpp0267478 | FBpp0267478 |
| FBpp0267102, FBpp0267103 | FBpp0267103 |
| FBpp0153606, FBpp0157663 | FBpp0157663 |
| FBpp0157330, FBpp0144755 | FBpp0144755 |
| FBpp0157205, FBpp0157374 | FBpp0157374 |
附录I 冗余序列的筛选列表
Appendix I Redundant sequences and the one finally selected
| 冗余序列 Redundant sequences | 所选的序列 Selected sequences |
|---|---|
| FBpp0073103, FBpp0073101, FBpp0073102 | FBpp0073102 |
| FBpp0078592, FBpp0110535, FBpp0078594, FBpp0078593 | FBpp0078593 |
| FBpp0084796, FBpp0084795 | FBpp0084795 |
| FBpp0110196, FBpp0078693 | FBpp0078693 |
| FBpp0087190, FBpp0111982, FBpp0111980, FBpp0111981 | FBpp0111981 |
| FBpp0086875, FBpp0086876 | FBpp0086876 |
| FBpp0087274, FBpp0087275, FBpp0087276 | FBpp0087276 |
| FBpp0083216, FBpp0083215 | FBpp0083215 |
| FBpp0099839, FBpp0099383 | FBpp0099383 |
| FBpp0268227, FBpp0264637 | FBpp0264637 |
| FBpp0259602, FBpp0267478 | FBpp0267478 |
| FBpp0267102, FBpp0267103 | FBpp0267103 |
| FBpp0153606, FBpp0157663 | FBpp0157663 |
| FBpp0157330, FBpp0144755 | FBpp0144755 |
| FBpp0157205, FBpp0157374 | FBpp0157374 |
| 重新注释的基因 Re-annotated genes | 对应的蛋白质 Corresponding proteins |
|---|---|
| Dsim\GD12690 | FBpp0211092 |
| Dmel\CG14102 | FBpp0074697 |
| Dyak\GE20089 | FBpp0265099 |
| Dpse\GA21694 | FBpp0280727 |
附录II 重新注释的基因
Appendix II Re-annotated genes
| 重新注释的基因 Re-annotated genes | 对应的蛋白质 Corresponding proteins |
|---|---|
| Dsim\GD12690 | FBpp0211092 |
| Dmel\CG14102 | FBpp0074697 |
| Dyak\GE20089 | FBpp0265099 |
| Dpse\GA21694 | FBpp0280727 |
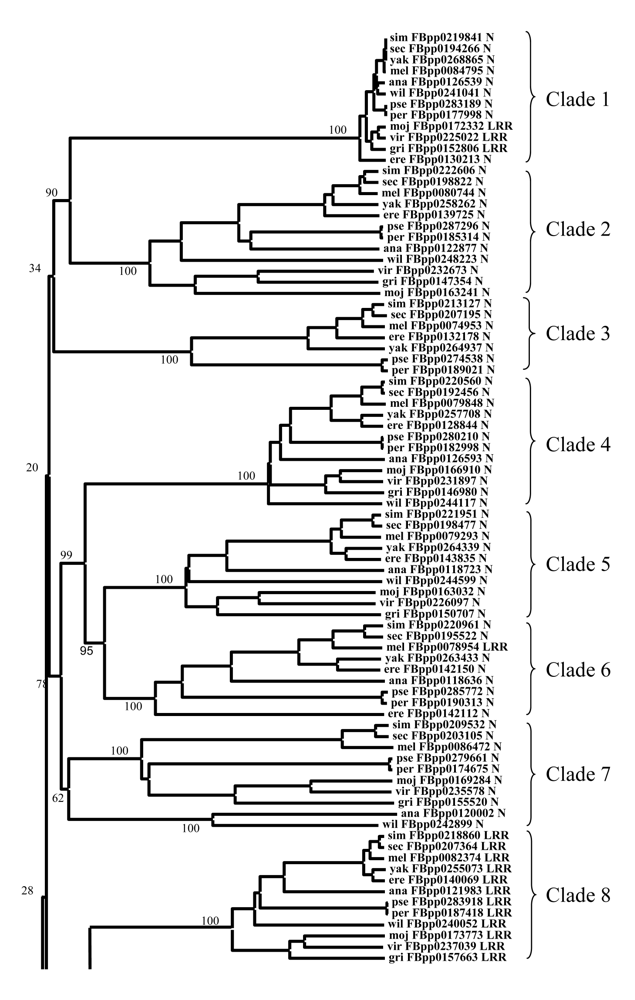
附录III 果蝇的12个近缘种中F-box蛋白质的系统发育关系。树中基因的名字用对应的物种的种加词前3个字母、蛋白质序列名和该蛋白质的C端结构域组成(其中N即none)。
Appendix III Phylogenetic tree of F-box proteins from 12 Drosophila species. The name of each protein is composed of the first three letters of the specific epithet, followed by the name of the sequence and the C-terminal domain (N means none).
| 进化枝编号 Clade no. | 合计 Total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 6 | 14 | 16 | 17 | 18 | 22 | 23 | 24 | 31 | 36 | 37 | 38 | ||
| 串联重复 Tandem duplication | 1 | 1 | 3 | 2 | 7 (29%) | |||||||||
| 散在重复 Dispersed duplication | 1 | 2 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 15 (63%) | ||||
| 反转录转座 Retroposition | 1 | 1 (4%) | ||||||||||||
| 从头起源 De novo origination | 1 | 1 (4%) | ||||||||||||
| 合计 Total | 1 | 2 | 1 | 1 | 7 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 2 | 24 (100%) |
附录IV 引起果蝇F-box基因拷贝数目增加的事件
Appendix IV Events caused increase in copy number of F-box genes in Drosophila
| 进化枝编号 Clade no. | 合计 Total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 6 | 14 | 16 | 17 | 18 | 22 | 23 | 24 | 31 | 36 | 37 | 38 | ||
| 串联重复 Tandem duplication | 1 | 1 | 3 | 2 | 7 (29%) | |||||||||
| 散在重复 Dispersed duplication | 1 | 2 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 15 (63%) | ||||
| 反转录转座 Retroposition | 1 | 1 (4%) | ||||||||||||
| 从头起源 De novo origination | 1 | 1 (4%) | ||||||||||||
| 合计 Total | 1 | 2 | 1 | 1 | 7 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 2 | 24 (100%) |

附录V D. mojavensis(a)和D. virilis(b)中的Clade18基因分别与D. willistoni和D. grimshawi中的非编码区序列的比对
Appendix V Sequence alignments of Clade18 genes from D. mojavensis and D. virilis with non-coding sequences from D. willistoni and D. grimshawi, respectively.
| 蛋白质名称 Protein name | C端结构域 C-terminal domain | 拷贝数变异 CNV | 参与的生物学过程 Biological process involved in |
|---|---|---|---|
| FBpp0073102 | WD40 | No | negative regulation of growth; regulation of mitosis; DNA endoreduplication. |
| FBpp0078846 | WD40 | No | protein ubiquitination during ubiquitin-dependent protein catabolic process; WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. |
| FBpp0083434 | WD40 | No | anatomical structure development; ovarian follicle cell development; gamete generation; regulation of biological process; circadian rhythm; learning or memory; cell motion; regulation of cellular component organization; olfactory learning; positive regulation of protein import into nucleus; locomotory behavior; catabolic process; negative regulation of nurse cell apoptosis; regulation of signal transduction; regulation of Wnt receptor signaling pathway. |
| FBpp0086876 | SPRY | No | negative regulation of synaptic growth at neuromuscular junction; neuromuscular synaptic transmission. |
| FBpp0077147 | UBCc | No | apoptosis; induction of compound eye retinal cell programmed cell death. |
| FBpp0078693 | LRR | No | circadian behavior; locomotor rhythm; entrainment of circadian clock by photoperiod. |
| FBpp0078970 | IBR | No | compound eye morphogenesis; negative regulation of protein catabolic process; G2 phase of mitotic cell cycle; G1/S transition of mitotic cell cycle; eye-antennal disc morphogenesis; regulation of mitosis. |
| FBpp0076199 | N | Yes | engulfment of apoptotic cell. |
| FBpp0073740 | LRR | Yes | deactivation of rhodopsin mediated signaling. |
附录VI F-box蛋白质的功能与拷贝数目变异的关系
Appendix VI Relationships between functions and copy number variation of F-box proteins
| 蛋白质名称 Protein name | C端结构域 C-terminal domain | 拷贝数变异 CNV | 参与的生物学过程 Biological process involved in |
|---|---|---|---|
| FBpp0073102 | WD40 | No | negative regulation of growth; regulation of mitosis; DNA endoreduplication. |
| FBpp0078846 | WD40 | No | protein ubiquitination during ubiquitin-dependent protein catabolic process; WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. |
| FBpp0083434 | WD40 | No | anatomical structure development; ovarian follicle cell development; gamete generation; regulation of biological process; circadian rhythm; learning or memory; cell motion; regulation of cellular component organization; olfactory learning; positive regulation of protein import into nucleus; locomotory behavior; catabolic process; negative regulation of nurse cell apoptosis; regulation of signal transduction; regulation of Wnt receptor signaling pathway. |
| FBpp0086876 | SPRY | No | negative regulation of synaptic growth at neuromuscular junction; neuromuscular synaptic transmission. |
| FBpp0077147 | UBCc | No | apoptosis; induction of compound eye retinal cell programmed cell death. |
| FBpp0078693 | LRR | No | circadian behavior; locomotor rhythm; entrainment of circadian clock by photoperiod. |
| FBpp0078970 | IBR | No | compound eye morphogenesis; negative regulation of protein catabolic process; G2 phase of mitotic cell cycle; G1/S transition of mitotic cell cycle; eye-antennal disc morphogenesis; regulation of mitosis. |
| FBpp0076199 | N | Yes | engulfment of apoptotic cell. |
| FBpp0073740 | LRR | Yes | deactivation of rhodopsin mediated signaling. |
| [1] |
Bai C, Richman R, Elledge SJ (1994) Human cyclin F. The EMBO Journal, 13,6087-6098.
URL PMID |
| [2] | Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Research, 30,276-280. |
| [3] |
Beckmann JS, Estivill X, Antonarakis SE (2007) Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nature Reviews Genetics, 8,639-646.
DOI URL PMID |
| [4] |
Cai J, Zhao R, Jiang H, Wang W (2008) De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics, 179,487-496.
DOI URL PMID |
| [5] |
Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, Hodgson A, George RA, Hoskins RA, Laverty T, Muzny DM, Nelson CR, Pacleb JM, Park S, Pfeiffer BD, Richards S, Sodergren EJ, Svirskas R, Tabor PE, Wan K, Stapleton M, Sutton GG, Venter C, Weinstock G, Scherer SE, Myers EW, Gibbs RA, Rubin GM (2002) Finishing a whole- genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biology, 3, research0079.1-0079.14.
DOI URL PMID |
| [6] |
Derti A, Roth FP, Church GM, Wu CT (2006) Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nature Genetics, 38,1216-1220.
DOI URL PMID |
| [7] |
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Developmental Cell, 9,109-119.
DOI URL PMID |
| [8] |
Drosophila 12 Genomes Consortium (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature, 450,203-218.
DOI URL PMID |
| [9] |
Eddy SR (1998) Profile hidden Markov models. Bioinformatics, 14,755-763.
DOI URL PMID |
| [10] |
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52,696-704.
DOI URL PMID |
| [11] |
Han J, Gon P, Reddig K, Mitra M, Guo P, Li HS (2006) The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell, 127,847-858.
URL PMID |
| [12] |
Hinds DA, Kloek AP, Jen M, Chen XY, Frazer KA (2006) Common deletions and SNPs are in linkage disequilibrium in the human genome. Nature Genetics, 38,82-85.
DOI URL PMID |
| [13] |
Junier T, Pagni M (2000) Dotlet: diagonal plots in a web browser. Bioinformatics, 16,178-179.
DOI URL PMID |
| [14] |
Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biology, 1,REVIEWS3002.
DOI URL PMID |
| [15] | Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ (2006) Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proceedings of the National Academy of Sciences, USA, 103,9935-9939. |
| [16] |
Li D, Dong Y, Jiang Y, Jiang H, Cai J, Wang W (2010) A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Research, 20,408-420.
DOI URL PMID |
| [17] |
Long M, Betran E, Thornton K, Wang W (2003) The origin of new genes: glimpses from the young and old. Nature Reviews Genetics, 4,865-875.
DOI URL PMID |
| [18] | Markow TA, O’Grady PM (2005) Drosophila: A Guide to Species Identification and Use, P. 250. Elsevier Academic, London. |
| [19] | Nei M (2007) The new mutation theory of phenotypic evolution. Proceedings of the National Academy of Sciences, USA, 104,12235-12242. |
| [20] | Nicholas K, Nicholas HB Jr (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the author. |
| [21] |
Niimura Y, Nei M (2006) Evolutionary dynamics of olfactory and other chemosensory receptor genes invertebrates. Journal of Human Genetics, 51,505-517.
URL PMID |
| [22] |
Niimura Y, Nei M (2007) Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One, 2,e708.
DOI URL PMID |
| [23] | Nozawa M, Nei M (2007) Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proceedings of the National Academy of Sciences, USA, 104,7122-7127. |
| [24] |
Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee C, Stone AC (2007) Diet and the evolution of human amylase gene copy number variation. Nature Genetics, 39,1256-1260.
DOI URL PMID |
| [25] |
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics, 14,817-818.
DOI URL PMID |
| [26] |
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen WW, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME (2006) Global variation in copy number in the human genome. Nature, 444,444-454.
URL PMID |
| [27] |
Reese MG, Hartze G, Harris NL, Ohler U, Abril JF, Lewis SE (2000) Genome annotation assessment in Drosophila melanogaster. Genome Research, 10,483-501.
DOI URL PMID |
| [28] |
Schaeffer SW, Bhutkar A, McAllister BF, Matsuda M, Matzkin LM, O’Grady PM, Rohde C, Valente VL, Aguadé M, Anderson WW, Edwards K, Garcia AC, Goodman J, Hartigan J, Kataoka E, Lapoint RT, Lozovsky ER, Machado CA, Noor MA, Papaceit M, Reed LK, Richards S, Rieger TT, Russo SM, Sato H, Segarra C, Smith DR, Smith TF, Strelets V, Tobari YN, Tomimura Y, Wasserman M, Watts T, Wilson R, Yoshida K, Markow TA, Gelbart WM, Kaufman TC (2008) Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics, 179,1601-1655.
DOI URL PMID |
| [29] |
Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE (2005) Segmental duplications and copy-number variation in the human genome. The American Journal of Human Genetics, 77,78-88.
DOI URL PMID |
| [30] |
Silva E, Au-Yeung HW, Van Goethem E, Burden J, Franc NC (2007) Requirement for a Drosophila E3-ubiquitin ligase in phagocytosis of apoptotic cells. Immunity, 27,585-596.
DOI URL PMID |
| [31] | Singh ND, Larracuente AM, Sackton TB, Clark AG (2009) Comparative genomics on the Drosophila phylogenetic tree. Annual Review of Ecology, Evolution, and Systematics, 40,459-480. |
| [32] | Small KS, Brudno M, Hill MM, Sidow A (2007) Extreme genomic variation in a natural population. Proceedings of the National Academy of Sciences, USA, 104,5698-5703. |
| [33] | Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, Ruby JG, Brennecke J, Harvard FlyBase curators, Berkeley Drosophila Genome Project, Hodges E, Hinrichs AS, Caspi A, Paten B, Park S, Han MV, Maeder ML, Polansky BJ, Robson BE, Aerts S, Helden J, Hassan B, Gilbert DG, Eastman DA, Rice M, Weir M, Hahn MW, Park Y, Pachter L, Kent WJ, Haussler D, Lai EC, Bartel DP, Hannon GJ, Kaufman TC, Eisen MB, Clark AG, Smith D, Celniker SE, Gelbart WM, Kellis M, Dewey CN (2007) Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature, 450,219-232. |
| [34] |
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24,1596-1599.
DOI URL PMID |
| [35] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25,4876-4882.
DOI URL PMID |
| [36] | Xu G, Ma H, Nei M, Kong H (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proceedings of the National Academy of Sciences, USA, 106,835-840. |
| [37] |
Zhou Q, Zhang G, Zhang Y, Xu S, Zhao R, Zhan Z, Li X, Ding Y, Yang S, Wang W (2008) On the origin of new genes in Drosophila. Genome Research, 18,1446-1455.
DOI URL PMID |
| [1] | 金泉泉, 向颖, 王华, 习新强. 南京仙林大学城三种绿地类型中果蝇多样性及其被寄生率[J]. 生物多样性, 2024, 32(8): 24156-. |
| [2] | 丁翔, 余元钧, 宋希强, 罗毅波. 具有泛化访花者的海芋特化传粉系统[J]. 生物多样性, 2024, 32(6): 24069-. |
| [3] | 公欣桐, 陈飞, 高欢欢, 习新强. 两种果蝇成虫与幼虫期的竞争及其对二者共存的影响[J]. 生物多样性, 2023, 31(8): 22603-. |
| [4] | 郭欣欣, 毛雪, 张敏. 锌对不同发育时期子代果蝇基因组DNA甲基化的影响[J]. 生物多样性, 2012, 20(6): 710-715. |
| [5] | 饶友生, 张细权. 拷贝数目变异: 基因组遗传变异的新诠释[J]. 生物多样性, 2008, 16(4): 399-406. |
| [6] | 钱远槐, 张文燕, 邓秋红, 张菁, 曾庆韬, 刘艳玲, 李守涛. 中国黑腹果蝇种组40种果蝇的核型多样性研究[J]. 生物多样性, 2006, 14(3): 188-205. |
| [7] | 郑向忠, 朱定良, 庚镇城, 张亚平. 果蝇Drosophila nasuta亚群系统发育问题的探讨[J]. 生物多样性, 1999, 07(3): 226-233. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn