



Biodiv Sci ›› 2011, Vol. 19 ›› Issue (3): 311-318. DOI: 10.3724/SP.J.1003.2011.08239 cstr: 32101.14.SP.J.1003.2011.08239
Special Issue: 物种形成与系统进化
Previous Articles Next Articles
Yan Liu1, Jianxiu Wang2, Xuejun Ge3,*( ), Tong Cao4,*(
), Tong Cao4,*( )
)
Received:2010-11-01
Accepted:2011-02-05
Online:2011-05-20
Published:2013-12-10
Contact:
Xuejun Ge,Tong Cao
Yan Liu, Jianxiu Wang, Xuejun Ge, Tong Cao. The rps4 locus as an alternative marker for barcoding bryophytes: eva- luation based on data mining from GenBank[J]. Biodiv Sci, 2011, 19(3): 311-318.
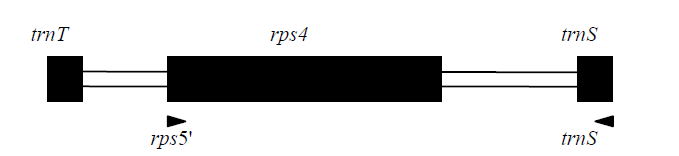
Fig. 1 The positions and directions of the primers used to amplify the rps4 cpDNA locus. Coding regions are highlighted by black boxes, whereas the non-coding regions are highlighted by white boxes.
| 种数 No. of species | 属数 No. of genera | 科数 No. of families | 目数 No. of orders | 序列数目 No. of accessions | |
|---|---|---|---|---|---|
| 藓纲 Bryophyta | 1,580 | 692 | 108 | 30 | 2,604 |
| 苔纲 Marchantiophyta | 498 | 184 | 74 | 15 | 756 |
| 角苔纲 Anthocerotophyta | 4 | 3 | 2 | 2 | 5 |
| 总计 Total | 2,082 | 879 | 184 | 47 | 3,365 |
Table 1 Characteristics of rps4 sequences from bryophytes available in GenBank
| 种数 No. of species | 属数 No. of genera | 科数 No. of families | 目数 No. of orders | 序列数目 No. of accessions | |
|---|---|---|---|---|---|
| 藓纲 Bryophyta | 1,580 | 692 | 108 | 30 | 2,604 |
| 苔纲 Marchantiophyta | 498 | 184 | 74 | 15 | 756 |
| 角苔纲 Anthocerotophyta | 4 | 3 | 2 | 2 | 5 |
| 总计 Total | 2,082 | 879 | 184 | 47 | 3,365 |
| 种内、种间遗传距离 数值的数目 Total no. of pair-wise comparisons | 物种识别率 Species resolution (%) | 无法识别的物种比例 Species unresolved (%) | |||
|---|---|---|---|---|---|
| 无种间变异 No inter-specific divergence (%) | 种内距离≥种间距离 Intra-specific distance ≥ inter-specific distance (%) | 种内变异> 0.2%* Intra-specific variations > 0.2%* (%) | |||
| 藓纲 Bryophyta | 19,664 | 70.7 (876/1,239) | 17.8 (221/1,239) | 7.6 (94/1,239) | 3.9 (48/1,239) |
| 苔纲 Marchantiophyta | 12,896 | 79.9 (333/417) | 9.6 (40/417) | 8.4 (35/417) | 2.2 (9/417) |
| 总计 Total | 3,2560 | 73.0 (1,209/1,656) | 15.8 (261/1,656) | 7.8 (129/1,656) | 3.4 (57/1,656) |
Table 2 Species resolution by the rps4 locus based on K2P distances
| 种内、种间遗传距离 数值的数目 Total no. of pair-wise comparisons | 物种识别率 Species resolution (%) | 无法识别的物种比例 Species unresolved (%) | |||
|---|---|---|---|---|---|
| 无种间变异 No inter-specific divergence (%) | 种内距离≥种间距离 Intra-specific distance ≥ inter-specific distance (%) | 种内变异> 0.2%* Intra-specific variations > 0.2%* (%) | |||
| 藓纲 Bryophyta | 19,664 | 70.7 (876/1,239) | 17.8 (221/1,239) | 7.6 (94/1,239) | 3.9 (48/1,239) |
| 苔纲 Marchantiophyta | 12,896 | 79.9 (333/417) | 9.6 (40/417) | 8.4 (35/417) | 2.2 (9/417) |
| 总计 Total | 3,2560 | 73.0 (1,209/1,656) | 15.8 (261/1,656) | 7.8 (129/1,656) | 3.4 (57/1,656) |
| 种数 No. of species | 序列数 No. of accessions | 物种识别率 Species resolution (%) | 种间无变异 No inter-specific divergence (%) | 种内距离≥种间距离 Intra-specific distance ≥ inter-specific distance (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rps4 | rbcL-a | rps4 | rbcL-a | rps4 | rbcL-a | rps4 | rbcL-a | rps4 | rbcL-a | |||||
| 羽苔属 Plagiochila | 128 | 111 | 147 | 125 | 82 | 77.5 | 8.6 | 20.7 | 9.4 | 1.8 | ||||
| 墙藓属 Tortula | 18 | 4 | 97 | 5 | 61.1 | - | 11.1 | - | 27.8 | - | ||||
| 匍灯藓属 Plagiomnium | 24 | 5 | 87 | 17 | 66.7 | 40 | 0 | 60 | 33.3 | 0 | ||||
| 桧藓属 Pyrrhobryum | 10 | 9 | 55 | 16 | 60 | 44.4 | 20 | 44.4 | 20 | 11.1 | ||||
| 小金发藓属 Pogonatum | 32 | 18 | 50 | 32 | 68.8 | 55.6 | 21.9 | 11.1 | 9.4 | 33.3 | ||||
| 紫萼藓属 Grimmia* | 35 | 29 | 49 | 71 | 71.4 | 58.6 | 22.9 | 20.7 | 5.7 | 20.7 | ||||
Table 3 Comparison of species resolution between rps4 and rbcL-a in six bryophyte genera
| 种数 No. of species | 序列数 No. of accessions | 物种识别率 Species resolution (%) | 种间无变异 No inter-specific divergence (%) | 种内距离≥种间距离 Intra-specific distance ≥ inter-specific distance (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rps4 | rbcL-a | rps4 | rbcL-a | rps4 | rbcL-a | rps4 | rbcL-a | rps4 | rbcL-a | |||||
| 羽苔属 Plagiochila | 128 | 111 | 147 | 125 | 82 | 77.5 | 8.6 | 20.7 | 9.4 | 1.8 | ||||
| 墙藓属 Tortula | 18 | 4 | 97 | 5 | 61.1 | - | 11.1 | - | 27.8 | - | ||||
| 匍灯藓属 Plagiomnium | 24 | 5 | 87 | 17 | 66.7 | 40 | 0 | 60 | 33.3 | 0 | ||||
| 桧藓属 Pyrrhobryum | 10 | 9 | 55 | 16 | 60 | 44.4 | 20 | 44.4 | 20 | 11.1 | ||||
| 小金发藓属 Pogonatum | 32 | 18 | 50 | 32 | 68.8 | 55.6 | 21.9 | 11.1 | 9.4 | 33.3 | ||||
| 紫萼藓属 Grimmia* | 35 | 29 | 49 | 71 | 71.4 | 58.6 | 22.9 | 20.7 | 5.7 | 20.7 | ||||
| [1] | CBOL Plant Working Group (2009) A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA, 106, 12794-12797. |
| [2] | Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñán S, Petersen G, Seberg O, Jørgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M (2007) A proposal for a standardised protocol to barcode all land plants. Taxon, 56, 295-299. |
| [3] |
Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Savolainen V (2005) Land plants and DNA barcodes: short-term and long-term goals. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 360, 1889-1895.
URL PMID |
| [4] |
Der JP, Thomson JA, Stratford JK, Wolf PG (2009) Global chloroplast phylogeny and biogeography of bracken (Pteridium; Dennstaedtiaceae). American Journal of Botany, 96, 1041-1049.
URL PMID |
| [5] | Devey DS, Chase MW, Clarkson JJ (2009) A stuttering start to plant DNA barcoding: microsatellites present a previously overlooked problem in non-coding plastid regions. Taxon, 58, 7-15. |
| [6] | Edwards D, Horn A, Taylor D, Savolainen V, Hawkins JA (2008) DNA barcoding of a large genus,Aspalathus L.(Fabaceae). Taxon, 57, 1317-1327. |
| [7] | Fahleson J, Okori P, Åkerblom-Espeby L, Dixelius C (2008) Genetic variability and genomic divergence of Elymus repens and related species. Plant Systematics and Evolution, 271, 143-156. |
| [8] |
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, Percy DM, Hajibabaei M, Barrett SCH (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE, 3, e2802. doi: 10.1371/journal.pone.0002802.
DOI URL PMID |
| [9] | Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, Newmaster SG, Hajibabaei M, Husband BC (2009) Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Molecular Ecology Resources, 9(s1), 130-139. |
| [10] | Gernandt DS, López GG, García SO, Liston A (2005) Phylogeny and classification of Pinus. Taxon, 54, 29-42. |
| [11] |
Goffinet B, Cox CJ, Shaw AJ, Hedderson TAJ (2001) The bryophyta (mosses): systematic and evolutionary inferences from an rps4 gene (cpDNA) phylogeny. Annals of Botany, 87, 191-208.
DOI URL PMID |
| [12] | Goffinet B, Shaw AJ (2009) Bryophyte Biology, 2nd edn. Cambridge University Press, Cambridge. |
| [13] |
Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA (2007) DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics, 23, 167-172.
URL PMID |
| [14] |
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Series B: Biological Sciences, 270, 313-321.
DOI URL PMID |
| [15] |
Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, Cowan R, Chase MW, Gaudeul M, Hollingsworth PM (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Molecular Ecology Resources, 9, 439-457.
URL PMID |
| [16] | Kelly LJ, Ameka GK, Chase MW (2010) DNA barcoding of African Podostemaceae (river-weeds): a test of proposed barcode regions. Taxon, 59, 251-260. |
| [17] |
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE, 2, e508. doi: 10.1371/journal.pone.0000508.
DOI URL PMID |
| [18] | Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences, USA, 102, 8369-8374. |
| [19] | Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. Proceedings of the National Academy of Sciences, USA, 105, 2923-2928. |
| [20] |
Li CX, Lu SG (2006) Phylogenetics of Chinese Dryopteris (Dryopteridaceae) based on the chloroplast rps4-trnS sequence data. Journal of Plant Research, 119, 589-598.
DOI URL PMID |
| [21] | Liu Y, Cao T, Ge XJ (2011) A case study of DNA barcoding in Chinese Grimmiaceae and a moss recorded in China for the first time. Taxon, 60, 185-193. |
| [22] |
Liu Y, Yan HF, Cao T, Ge XJ (2010) Evaluation of 10 plant barcodes in Bryophyta (Mosses). Journal of Systematics and Evolution, 48, 36-46.
DOI URL |
| [23] | Nadot S, Bajon R, Lejeune B (1994) The chloroplast gene rps4 as a tool for the study of Poaceae phylogeny. Plant Systematics and Evolution, 191, 27-38. |
| [24] | Newmaster SG, Fazekas AJ, Ragupathy S (2006) DNA barcoding in the land plants: evaluation of rbcL in a multigene tiered approach. Canadian Journal of Botany, 84, 335-341. |
| [25] | Newmaster SG, Ragupathy S (2009) Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Molecular Ecology Resources, 9(s1), 172-180. |
| [26] |
Pennisi E (2007) Wanted: a barcode for plants. Science, 318, 190-191.
DOI URL PMID |
| [27] | Ragupathy S, Newmaster SG, Murugesan M, Balasubramaniam V (2009) DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the hill tribes of the Western Ghats in southern India. Molecular Ecology Resources, 9(s1), 164-171. |
| [28] |
Seberg O, Petersen G (2009) How many loci does it take to DNA barcode a Crocus? PLoS ONE, 4, e4598. doi: 10.1371/journal.pone.0004598.
DOI URL PMID |
| [29] | Shaw AJ (2001) Biogeographic patterns and cryptic speciation in bryophytes. Journal of Biogeography, 28, 253-261. |
| [30] | Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B (1997) Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Systematics and Evolution, 204, 109-123. |
| [31] |
Steele PR, Friar LM, Gilbert LE, Jansen RK (2010) Molecular systematics of the neotropical genus Psiguria (Cucurbitaceae): implications for phylogeny and species identification. American Journal of Botany, 97, 156-173.
URL PMID |
| [32] |
Stenøien HK (2008) Slow molecular evolution in 18S rDNA,rbcL and nad5 genes of mosses compared with higher plants. Journal of Evolutionary Biology, 21, 566-571.
DOI URL PMID |
| [33] | Swofford DL (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0 b10. Sinauer Associates, Sunderland. |
| [34] |
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Research, 35, e14.
URL PMID |
| [35] |
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599.
URL PMID |
| [36] |
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI URL PMID |
| [37] | Vitt DH, Wieder RK (2009) The structure and function of bryophyte-dominated peatlands. In: Bryophyte Biology, 2nd edn. (eds Goffinet B, Shaw AJ), pp. 357-392. Cambridge University Press, Cambridge. |
| [38] |
Werner O, Guerra J (2004) Molecular phylogeography of the moss Tortula muralis Hedw. (Pottiaceae) based on chloroplast rps4 gene sequence data. Plant Biology, 6, 147-157.
DOI URL PMID |
| [39] | Werner O, Ros RM, Cano MJ, Guerra J (2004) Molecular phylogeny of Pottiaceae (Musci) based on chloroplast rps4 sequence data. Plant Systematics and Evolution, 243, 147-164. |
| [40] | Wyatt R, Odrzykoski IJ (1998) On the origins of the allopolyploid moss Plagiomnium cuspidatum. Bryologist, 101, 263-271. |
| [41] | Wyatt R, Odrzykoski IJ, Stoneburner A (1992) Isozyme evidence of reticulate evolution in mosses:Plagiomnium medium is an allopolyploid of P. ellipticum × P. insigne. Systematic Botany, 17, 532-550. |
| [42] | Wyatt R, Odrzykoski IJ, Stoneburner A (1993) Isozyme evidence regarding the origins of the allopolyploid moss Plagiomnium curvatulum. Lindbergia, 18, 49-58. |
| [43] | Wyatt R, Odrzykoski IJ, Stoneburner A, Bass HW, Galau GA (1988) Allopolyploidy in bryophytes: multiple origins of Plagiomnium medium. Proceedings of the National Academy of Sciences, USA, 85, 5601-5604. |
| [44] | Zander RH (1993) Genera of Pottiaceae: Mosses of Harsh Environments. Buffalo Society of Natural Sciences, Buffalo. |
| [1] | Keyi Wu, Wenda Ruan, Difeng Zhou, Qingchen Chen, Chengyun Zhang, Xinyuan Pan, Shang Yu, Yang Liu, Rongbo Xiao. Syllable clustering analysis-based passive acoustic monitoring technology and its application in bird monitoring [J]. Biodiv Sci, 2023, 31(1): 22370-. |
| [2] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [3] | Jiaxin Kong, Zhaochen Zhang, Jian Zhang. Classification and identification of plant species based on multi-source remote sensing data: Research progress and prospect [J]. Biodiv Sci, 2019, 27(7): 796-812. |
| [4] | Yun Cao, Wenjing Shen, Lian Chen, Feilong Hu, Lei Zhou, Haigen Xu. Application of metabarcoding technology in studies of fungal diversity [J]. Biodiv Sci, 2016, 24(8): 932-939. |
| [5] | Tai Wang, Yanping Zhang, Lihong Guan, Yanyan Du, Zhongyu Lou, Wenlong Jiao. Current freshwater fish resources and the application of DNA barcoding in species identification in Gansu Province [J]. Biodiv Sci, 2015, 23(3): 306-313. |
| [6] | Dangni Zhang, Lianming Zheng, Jinru He, Wenjing Zhang, Yuanshao Lin, Yang Li. DNA barcoding of hydromedusae in northern Beibu Gulf for species identification [J]. Biodiv Sci, 2015, 23(1): 50-60. |
| [7] | Chaolun Li, Minxiao Wang, Fangping Cheng, Song Sun. DNA barcoding and its application to marine zooplankton ecology [J]. Biodiv Sci, 2011, 19(6): 805-814. |
| [8] | Haifeng Gu, Tingting Liu, Dongzhao Lan. Progress of dinoflagellate cyst research in the China seas [J]. Biodiv Sci, 2011, 19(6): 779-786. |
| [9] | Yiyi Long, Liyuan Yang, Wanjin Liao. Identifying cryptic species in pollinating-fig wasps by PCR-RFLP on mtDNA COI gene [J]. Biodiv Sci, 2010, 18(4): 414-419. |
| [10] | Shuping Ning, Haifei Yan, Gang Hao, Xuejun Ge. Current advances of DNA barcoding study in plants [J]. Biodiv Sci, 2008, 16(5): 417-425. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()