



Biodiv Sci ›› 2018, Vol. 26 ›› Issue (12): 1289-1295. DOI: 10.17520/biods.2018121 cstr: 32101.14.biods.2018121
• Original Papers: Animal Diversity • Previous Articles Next Articles
Jiping Yang1, Ce Li1,2, Weitao Chen1, Yuefei Li1, Xinhui Li1,*( )
)
Received:2018-04-16
Accepted:2018-04-16
Online:2018-12-20
Published:2019-02-11
Contact:
Li Xinhui
About author:# 同等贡献作者 Contributed equally to this work
Jiping Yang, Ce Li, Weitao Chen, Yuefei Li, Xinhui Li. Genetic diversity and population demographic history of Ochetobius elongatus in the middle and lower reaches of the Xijiang River[J]. Biodiv Sci, 2018, 26(12): 1289-1295.
| 采样点 Sample site | 采样日期 Sampling time | 样本量 Sample size | 经度 Longitude | 纬度 Latitude |
|---|---|---|---|---|
| 广西桂平 Guiping | 2017.09 | 1 | 110°05°54.78°° E | 23°24°08.25°° N |
| 广西梧州 Wuzhou | 2017.09 | 1 | 111°17°15.32°° E | 23°28°07.94°° N |
| 广东肇庆 Zhaoqing | 2011.05 | 22 | 112°23°40.13°° E | 23°05°06.24°° N |
| 广东肇庆 Zhaoqing | 2013.05 | 28 | 112°23°40.13°° E | 23°05°06.24°° N |
Table 1 Sample information of the present study
| 采样点 Sample site | 采样日期 Sampling time | 样本量 Sample size | 经度 Longitude | 纬度 Latitude |
|---|---|---|---|---|
| 广西桂平 Guiping | 2017.09 | 1 | 110°05°54.78°° E | 23°24°08.25°° N |
| 广西梧州 Wuzhou | 2017.09 | 1 | 111°17°15.32°° E | 23°28°07.94°° N |
| 广东肇庆 Zhaoqing | 2011.05 | 22 | 112°23°40.13°° E | 23°05°06.24°° N |
| 广东肇庆 Zhaoqing | 2013.05 | 28 | 112°23°40.13°° E | 23°05°06.24°° N |
| 引物名 Primer | 引物序列 Primer sequence (5′-3′) | 退火温度 Annealed temperature (℃) | 参考文献 Reference | |
|---|---|---|---|---|
| Cytb | L14724 | GACTTGAAA AACCACCGTTG | 58-64 | Xiao et al, 2001 |
| H15915 | CTCCGATCTCCGGATTACAAGAC | |||
| ND2 | AFND2L | AAGCTYTYGGGCCCATACC | 58 | 黎瑞宝, 2013① |
| AFND2R | TCCYGCTTAGGGCTTTGAAGG | |||
| myh6 | myh6_F459 | CATMTTYTCCATCTCAGATAATGC | 53 | Chen et al, 2008 |
| myh6_R1325 | ATTCTCACCACCATCCAGTTGAA | |||
| RAG2 | RAG2-f2a | AARCGCTCMTGTCCMACTGG | 55 | Lovejoy & Collette, 2001 |
| RAG2-R6a | TGRTCCARGCAGAAGTACTTG |
Table 2 Primer information used in the present study
| 引物名 Primer | 引物序列 Primer sequence (5′-3′) | 退火温度 Annealed temperature (℃) | 参考文献 Reference | |
|---|---|---|---|---|
| Cytb | L14724 | GACTTGAAA AACCACCGTTG | 58-64 | Xiao et al, 2001 |
| H15915 | CTCCGATCTCCGGATTACAAGAC | |||
| ND2 | AFND2L | AAGCTYTYGGGCCCATACC | 58 | 黎瑞宝, 2013① |
| AFND2R | TCCYGCTTAGGGCTTTGAAGG | |||
| myh6 | myh6_F459 | CATMTTYTCCATCTCAGATAATGC | 53 | Chen et al, 2008 |
| myh6_R1325 | ATTCTCACCACCATCCAGTTGAA | |||
| RAG2 | RAG2-f2a | AARCGCTCMTGTCCMACTGG | 55 | Lovejoy & Collette, 2001 |
| RAG2-R6a | TGRTCCARGCAGAAGTACTTG |
| 单倍型数/个体数 Number of haplotype/ sample size | 单倍型多态性 Haplotype diversity | 近期核苷酸多态性 Current nucleotide diversity (θπ, %) | 历史核苷酸多态性 Historical nucleotide diversity (θw, %) | Tajima’s D 检验 Tajima’s D test | Fu’s Fs检验 Fu’s Fs test | 最优模型 Optimal model | |
|---|---|---|---|---|---|---|---|
| Cytb | 13/52 | 0.784 | 0.197 | 0.470 | -1.83** | -5.43** | HKY+I |
| ND2 | 17/52 | 0.869 | 0.239 | 0.272 | -0.35 | -8.16*** | F81+I+G |
| Cytb + ND2 | 26/52 | 0.940 | 0.219 | 0.366 | -1.33 | -14.54*** | - |
| myh6 | 2/52 | 0.462 | 0.060 | 0.029 | - | - | HKY |
| RAG2 | 3/52 | 0.147 | 0.022 | 0.077 | - | - | F81 |
| myh6 + RAG2 | 5/52 | 0.546 | 0.040 | 0.055 | - | - | - |
Table 3 Genetic diversity indices, neutrality tests and optimal nucleotide substitution model of Ochetobius elongatus
| 单倍型数/个体数 Number of haplotype/ sample size | 单倍型多态性 Haplotype diversity | 近期核苷酸多态性 Current nucleotide diversity (θπ, %) | 历史核苷酸多态性 Historical nucleotide diversity (θw, %) | Tajima’s D 检验 Tajima’s D test | Fu’s Fs检验 Fu’s Fs test | 最优模型 Optimal model | |
|---|---|---|---|---|---|---|---|
| Cytb | 13/52 | 0.784 | 0.197 | 0.470 | -1.83** | -5.43** | HKY+I |
| ND2 | 17/52 | 0.869 | 0.239 | 0.272 | -0.35 | -8.16*** | F81+I+G |
| Cytb + ND2 | 26/52 | 0.940 | 0.219 | 0.366 | -1.33 | -14.54*** | - |
| myh6 | 2/52 | 0.462 | 0.060 | 0.029 | - | - | HKY |
| RAG2 | 3/52 | 0.147 | 0.022 | 0.077 | - | - | F81 |
| myh6 + RAG2 | 5/52 | 0.546 | 0.040 | 0.055 | - | - | - |
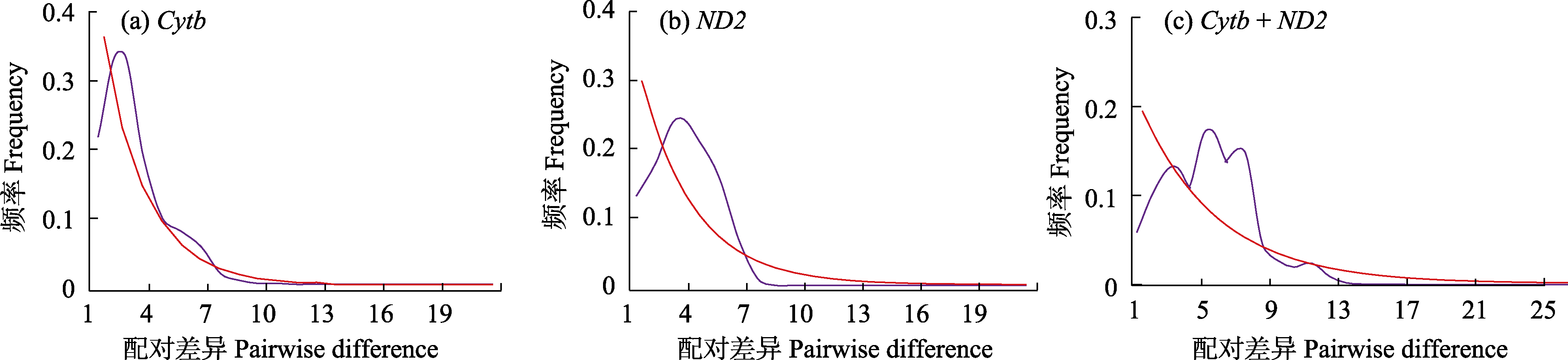
Fig. 2 Mismatch distribution analysis based on different genes and gene groups. Red and purple lines represent observed value and expected values, respectively.
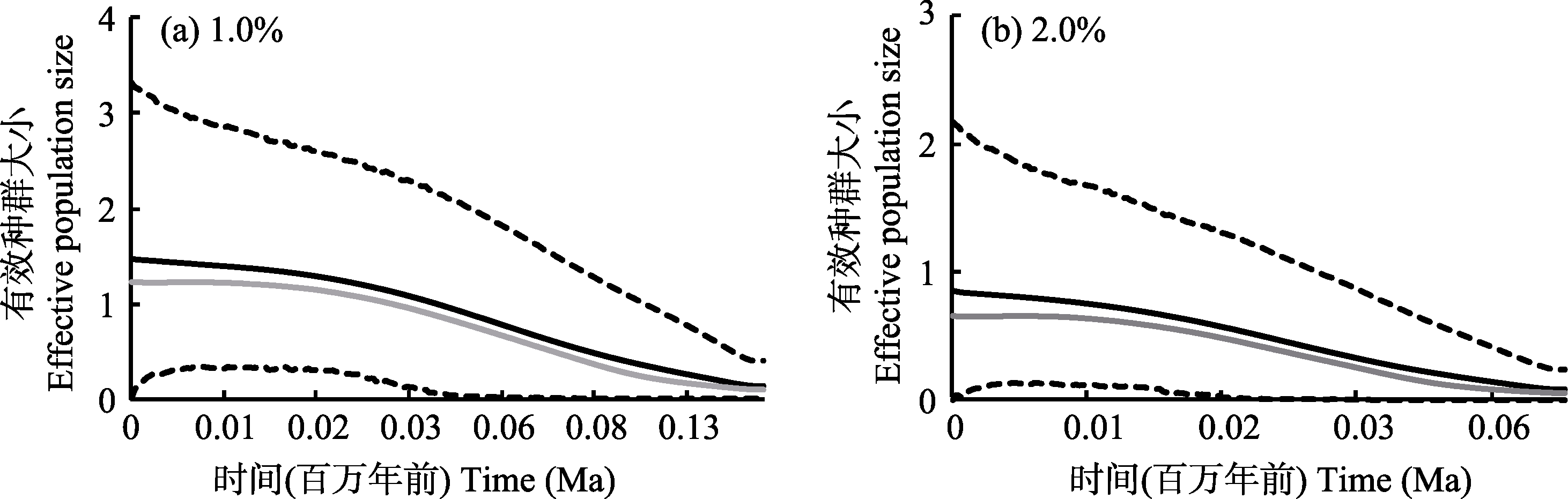
Fig. 3 Extended Bayesian Skyline plots. Black lines represent mean values of effective population size, gray lines represent median values of effective population size, and dashed lines represent 95% confidence interval of effective population size.
| 1 | Araujo-Lima CARM, Oliveira EC (1998) Transport of larval fish in the Amazon. Journal of Fish Biology, 53, 297-306. |
| 2 | Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| 3 | Chen WJ, Miya M, Saitoh K, Mayden RL (2008) Phylogenetic utility of two existing and four novel nuclear gene loci in reconstructing Tree of Life of ray-finned fishes: The order Cypriniformes (Ostariophysi) as a case study. Gene, 423, 125-134. |
| 4 | Chen YY, Cao WX, Zheng CY (1986) Ichthyofauna of the Zhujiang River with a discussion on zoogeographical divisions for freshwater fishes. Acta Hydrobiologica Sinica, 10, 228-234. (in Chinese with English abstract) |
| [陈宜瑜, 曹文宣, 郑慈英 (1986) 珠江的鱼类区系及其动物地理区划的讨论. 水生生物学报, 10, 228-234.] | |
| 5 | Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214. |
| 6 | Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution, 22, 1185-1192. |
| 7 | Durand JD, Tsigenopoulos CS, Unlu E, Berrebi P (2002) Phylogeny and biogeography of the family Cyprinidae in the Middle East inferred from cytochrome b DNA—Evolutionary significance of this region. Molecular Phylogenetics and Evolution, 25, 91-100. |
| 8 | Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792-1797. |
| 9 | Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567. |
| 10 | Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915-925. |
| 11 | Gascoyne M, Benjamin GJ, Schwarcz HP, Ford DC (1979) Sea-level lowering during the Illinoian glaciation: Evidence from a Bahama “blue hole”. Science, 205, 806-808. |
| 12 | Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426. |
| 13 | Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B-Biological Sciences, 359, 183-195. |
| 14 | Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng GM, Dong L, Zhang ZW, Ding P, Luo ZH, Ding CQ, Ma ZJ, Tang SH, Cao WX, Li CW, Hu HJ, Ma Y, Wu Y, Wang YX, Zhou KY, Liu SY, Chen YY, Li JT, Feng ZJ, Wang Y, Wang B, Li C, Song XL, Cai L, Zang CX, Zeng Y, Meng ZB, Fang HX, Ping XG (2016) Red List of China’s Vertebrates. Biodiversidy Science, 24, 500-551. |
| [蒋志刚, 江建平, 王跃招, 张鹗, 张雁云, 李立立, 谢锋, 蔡波, 曹亮, 郑光美, 董路, 张正旺, 丁平, 罗振华, 丁长青, 马志军, 汤宋华, 曹文宣, 李春旺, 胡慧建, 马勇, 吴毅, 王应祥, 周开亚, 刘少英, 陈跃英, 李家堂冯祚建, 王燕, 王斌, 李成, 宋雪琳, 蔡蕾, 臧春鑫, 曾岩, 孟智斌, 方红霞, 平晓鸽 (2016) 中国脊椎动物红色名录. 生物多样性, 24, 500-551.] | |
| 15 | Ketmaier V, Bianco PG, Cobollia M, Krivokapic M, Caniglia R, De Matthaeis E (2004) Molecular phylogeny of two lineages of Leuciscinae cyprinids (Telestes and Scardinius) from the peri-Mediterranean area based on cytochrome b data. Molecular Phylogenetics and Evolution, 32, 1061-1071. |
| 16 | Li J, Li XH, Jia XP, Li YF, He MF, Tan XC, Wang C, Jiang WX (2010) Evolvement and diversity of fish community in Xijiang River. Journal of Fishery Sciences of China, 17, 298-311. (in Chinese with English abstract) |
| [李捷, 李新辉, 贾晓平, 李跃飞, 何美峰, 谭细畅, 王超, 蒋万祥 (2010) 西江鱼类群落多样性及其演变. 中国水产科学, 17, 298-311.] | |
| 17 | Li SF, Lü GQ, Bernatchez L (1998) Diversity of mitochondrial DNA in the populations of silver carp, bighead carp, grass carp and black carp in the middle- and lower reaches of the Yangtze River. Acta Zoologica Sinica, 44, 82-93. (in Chinese with English abstract) |
| [李思发, 吕国庆, Bernatchez L (1998) 长江中下游鲢鳙草青四大家鱼线粒体DNA多样性分析. 动物学报, 44, 82-93.] | |
| 18 | Li YF, Li XH, Tan XC, Li J, Wang C (2012) Occurrence of larval Elopichthys bambusa and its relationship with hydrological conditions in the middle and lower reaches of Pearl River. Journal of Fisheries of China, 36(4), 15-22. (in Chinese with English abstract) |
| [李跃飞, 李新辉, 谭细畅, 李捷, 王超 (2012) 珠江中下游鳡鱼苗的发生及其与水文环境的关系. 水产学报, 36, 15-22.] | |
| 19 | Liang ZS, Mo RL, Chen FC (1985) Species identification and spawning type of common fish species in Xijiang River during early phase. In: The Report of Fishery Research in Pearl River Basin. Compilation Committee of Fishery Resources Survey of Pearl River Basin, Guangzhou. (in Chinese) |
| [梁秩燊, 莫瑞林, 陈福才 (1985) 西江常见鱼类早期发育的分类鉴定及其产卵类型. 见: 珠江水系渔业资源调查研究报告. 珠江水系渔业资源调查编委会, 广州.] | |
| 20 | Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452. |
| 21 | Lovejoy NR, Collette BB (2001) Phylogenetic relationships of new world needlefishes (Teleostei: Belonidae) and the biogeography of transitions between marine and freshwater habitats. Copeia, 2001, 324-338. |
| 22 | Meyer A (1993) Evolution of Mitochondrial DNA in Fishes. Elsevier, The Hague. |
| 23 | Nesler TP, Muth RT, Wasowicz AF (1988) Evidence for baseline flow spikes as spawning cues for Colorado squawfish in the Yampa River, Colorado. In: American Fisheries Society Symposium, pp. 68-79. Bethesda, Maryland. |
| 24 | Nylander JAA (2004) MrModeltest v2. Evolutionary Biology Centre, Uppsala University, Uppsala. |
| 25 | Rambaut A, Drummond A (2007) Tracer v1.4.. (accessed on 2018-3-2). |
| 26 | Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569. |
| 27 | Tajima F (1989) The effect of change in population size on DNA polymorphism. Genetics, 123, 597-601. |
| 28 | Tao W, Zou M, Wang X, Gan X, Mayden RL, He S (2010) Phylogenomic analysis resolves the formerly intractable adaptive diversification of the endemic clade of east Asian Cyprinidae (Cypriniformes). PLoS ONE, 5, e13508. |
| 29 | Tyus HM, Haines GB (1991) Distribution, habitat use, and growth of age-0 Colorado squawfish in the Green River basin, Colorado and Utah. Transactions of the American Fisheries Society, 120, 79-89. |
| 30 | Wang X, Li J, He S (2007) Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Molecular Phylogenetics and Evolution, 42, 157-170. |
| 31 | Wu WJ, Peng M, Wang DP, Shi J, Li YS, Han YQ, Lei JJ, He AY (2015) Comparison of mitochondrial D-Loop and Cyt b sequences of Hypophthalmichthys molitrix. Open Journal of Fisheries Research, 2(4), 67-73. (in Chinese with English abstract) |
| [吴伟军, 彭敏, 王大鹏, 施军, 李育森, 韩耀全, 雷建军, 何安尤 (2015) 红水河鲢线粒体D-Loop和Cyt b基因序列分析. 水产研究, 2(4), 67-73.] | |
| 32 | Xiao W, Zhang Y, Liu H (2001) Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Molecular Phylogenetics and Evolution, 18, 163-173. |
| 33 | Yue PQ (2000) Fauna Sinica, Osteichthyes, Cypriniformes II Science Press, Beijing. (in Chinese) |
| [乐佩琦 (2000) 中国动物志. 硬骨鱼纲, 鲤形目, 中卷. 科学出版社, 北京.] | |
| 34 | Zheng CY (1989) The Ichthyography of the Pearl River. Science Press, Beijing. (in Chinese) |
| [郑慈英 (1989) 珠江鱼类志. 科学出版社. 北京.] | |
| 35 | Zitek A, Schmutz S, Unfer G, Ploner A (2004) Fish drift in a Danube sidearm-system: I. Site-, inter- and intraspecific patterns. Journal of Fish Biology, 65, 1319-1338. |
| [1] | Jing Gan Xiangxu Liu Xueming Lu Xing Yue. China's Large Cities in Global Biodiversity Hotspots: Conservation Policies and Optimization Directions [J]. Biodiv Sci, 2025, 33(5): 24529-. |
| [2] | Zixuan Zeng Rui Yang Yue Huang Luyao Chen. Characteristics of bird diversity and environmental relationships in Tsinghua University campus [J]. Biodiv Sci, 2025, 33(5): 24373-. |
| [3] | Hao Zhou, Mingyi Wang, Chuge Zhang, Zhishu Xiao, Fang Ouyang. The status and challenges of insect hotels in the conservation of urban solitary bees and wasps diversity [J]. Biodiv Sci, 2025, 33(5): 24472-. |
| [4] | Xiaoyu Zhu, Chenhao Wang, Zhongjun Wang, Yujun Zhang. Research progress and prospect of urban green space biodiversity [J]. Biodiv Sci, 2025, 33(5): 25027-. |
| [5] | Xin Wang, Femgyu Bao. Analysis of the ecological restoration effect of South Dianchi National Wetland Park based on the enhancement of bird diversity [J]. Biodiv Sci, 2025, 33(5): 24531-. |
| [6] | Yue Ming, Peiyao Hao, Lingqian Tan, Xi Zheng. A study on urban biodiversity conservation and enhancement in china based on the concept of green and high-quality development of cities [J]. Biodiv Sci, 2025, 33(5): 24524-. |
| [7] | Murong Yi, Ping Lu, Yong Peng, Yong Tang, Jiuheng Xu, Haoping Yin, Luyang Zhang, Xiaodong Weng, Mingxiao Di, Juan Lei, Chenqi Lu, Rujun Cao, Nianhua Dai, Deyang Zhan, Mei Tong, Zhiming Lou, Yonggang Ding, Jing Chai, Jing Che. Population status and habitat of Critically Endangered Jiangxi giant salamander (Andrias jiangxiensis) [J]. Biodiv Sci, 2025, 33(4): 24145-. |
| [8] | Motong Li, Tuo He, Wei Li, Jing Liao, Yan Zeng. Regulating international trade in wild fauna and flora: An analysis of CITES terminology [J]. Biodiv Sci, 2025, 33(4): 24545-. |
| [9] | Xiaoqiang Lu, Shanshan Dong, Yue Ma, Xu Xu, Feng Qiu, Mingyue Zang, Yaqiong Wan, Luanxin Li, Cigang Yu, Yan Liu. Current status, challenges, and prospects of frontier technologies in biodiversity conservation applications [J]. Biodiv Sci, 2025, 33(4): 24440-. |
| [10] | Guo Yutong, Li Sucui, Wang Zhi, Xie Yan, Yang Xue, Zhou Guangjin, You Chunhe, Zhu Saning, Gao Jixi. Coverage and distribution of national key protected wild species in China’s nature reserves [J]. Biodiv Sci, 2025, 33(3): 24423-. |
| [11] | Zhao Weiyang, Wang Wei, Ma Bingran. Advances and prospects in research on other effective area-based conservation measures (OECMs) [J]. Biodiv Sci, 2025, 33(3): 24525-. |
| [12] | Zhou Zhihua, Jin Xiaohua, Luo Ying, Li Diqiang, Yue Jianbing, Liu Fang, He Tuo, Li Xi, Dong Hui, Luo Peng. Analyses and suggestions on mechanisms of forestry and grassland administrations in China to achieve targets of Kunming-Montreal Global Biodiversity Framework [J]. Biodiv Sci, 2025, 33(3): 24487-. |
| [13] | Liu Li, Zang Mingyue, Ma Yue, Wan Yaqiong, Hu Feilong, Lu Xiaoqiang, Liu Yan. Measures, progress and prospects of central-local cooperation in the implementation of the National Biodiversity Strategy and Action Plan [J]. Biodiv Sci, 2025, 33(3): 24532-. |
| [14] | Yang Song, Jun Liu, Shaolin He, Wei Xu, Chen Cheng, Bo Liu, Jiqing Yu. Mainstreaming path of biodiversity conservation for Chinese energy enterprises [J]. Biodiv Sci, 2025, 33(1): 24345-. |
| [15] | Tuo He, Yan Zeng, Yafang Yin, Kun Zhang, Liangchen Yuan, Hui Dong, Zhihua Zhou. Consolidating the scientific foundation for global wild plant conservation and sustainable trade—Comments on the 27th Meeting of the Plants Committee of CITES [J]. Biodiv Sci, 2024, 32(9): 24390-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()