



生物多样性 ›› 2019, Vol. 27 ›› Issue (11): 1172-1183. DOI: 10.17520/biods.2019113 cstr: 32101.14.biods.2019113
所属专题: 物种形成与系统进化
收稿日期:2019-04-02
接受日期:2019-07-07
出版日期:2019-11-20
发布日期:2019-11-20
通讯作者:
卢金梅
基金资助:
Xinyu Du1,2,Jinmei Lu1,*( ),Dezhu Li1,2
),Dezhu Li1,2
Received:2019-04-02
Accepted:2019-07-07
Online:2019-11-20
Published:2019-11-20
Contact:
Lu Jinmei
摘要:
近年来, 随着测序技术的发展, 石松类和蕨类植物的核基因组、质体基因组以及线粒体基因组研究发展迅速, 质体基因组研究工作更是呈爆发式增长。截至2019年3月1日, GenBank公布的石松类和蕨类植物的175个质体基因组中, 约3/4为最近两年新增。研究内容从早期对个别质体基因组结构和序列特征的简单报道, 逐渐发展到综合性的比较基因组学和系统发育基因组学研究。目前已发表的质体基因组覆盖了石松类和蕨类植物的所有目和大部分科, 这两大类群的质体基因组结构变异和系统发育的基本框架已逐渐清晰。这些研究为我们理解维管植物的早期演化提供了重要参考。本文对石松类和蕨类植物的质体基因组结构特征进行了系统梳理, 发现其结构变异主要包括大片段倒位、IR区边界变动、基因或内含子丢失等, 其中一些结构变异可作为较高分类阶元的共衍征。RNA编辑和长片段非编码序列插入普遍存在于石松类和蕨类植物的质体基因组中, 但其起源、演化机制和功能等仍不清楚。我们对质体基因组的应用、系统发育研究中质体和核基因组的优劣性, 以及系统发育基因组学的前景进行了评述。
杜新宇, 卢金梅, 李德铢 (2019) 石松类和蕨类植物质体基因组结构演化研究进展. 生物多样性, 27, 1172-1183. DOI: 10.17520/biods.2019113.
Xinyu Du, Jinmei Lu, Dezhu Li (2019) Advances in the evolution of plastid genome structure in lycophytes and ferns. Biodiversity Science, 27, 1172-1183. DOI: 10.17520/biods.2019113.
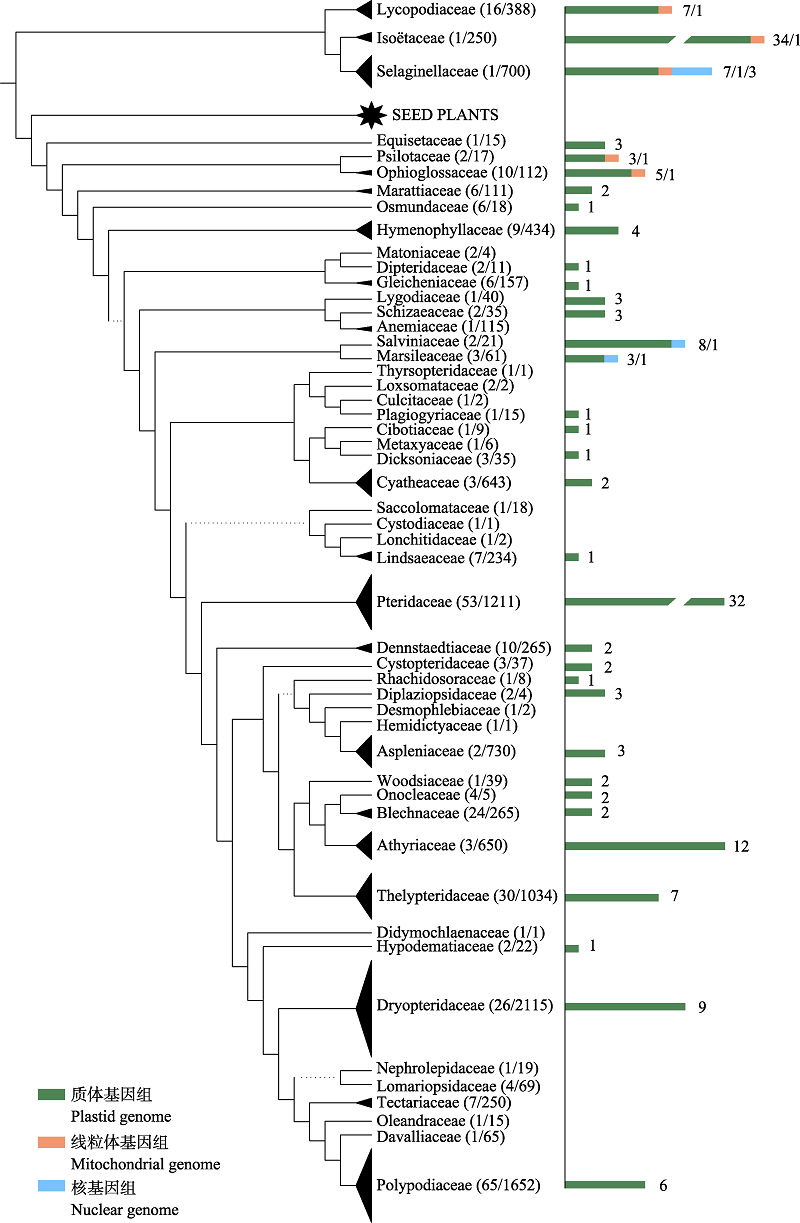
图1 石松类和蕨类植物质体、线粒体和核基因组已发表情况。以科为单位统计,用柱状图和数字标注在系统树右侧。系统树基于 PPG I (2016)修改。
Fig. 1 The number of published plastid, mitochondrial and nuclear genomes for lycophytes and ferns. The numbers were measured in families and were indicated to the right of the phylogenetic tree with histograms and numbers. The phylogenetic tree is modified from PPG I (2016).
| 1 | Aya K, Kobayashi M, Tanaka J, Ohyanagi H, Suzuki T, Yano Kenji, Takano T, Yano K, Matsuoka M ( 2015) De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database, Ljtrans DB. Plant and Cell Physiology, 56, e5. |
| 2 | Bakker FT ( 2017) Herbarium genomics: Skimming and plastomics from archival specimens. Webbia, 72, 1-11. |
| 3 | Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, Ashton NW, Axtell MJ, Barker E, Barker MS, Bennetzen JL, Bonawitz ND, Chapple C, Cheng C, Correa LG, Dacre M, DeBarry J, Dreyer I, Elias M, Engstrom EM, Estelle M, Feng L, Finet C, Floyd SK, Frommer WB, Fujita T, Gramzow L, Gutensohn M, Harholt J, Hattori M, Heyl A, Hirai T, Hiwatashi Y, Ishikawa M, Iwata M, Karol KG, Koehler B, Kolukisaoglu U, Kubo M, Kurata T, Lalonde S, Li K, Li Y, Litt A, Lyons E, Manning G, Maruyama T, Michael TP, Mikami K, Miyazaki S, Morinaga S, Murata T, Mueller-Roeber B, Nelson DR, Obara M, Oguri Y, Olmstead RG, Onodera N, Petersen BL, Pils B, Prigge M, Rensing SA, Riano-Pachon DM, Roberts AW, Sato Y, Scheller HV, Schulz B, Schulz C, Shakirov EV, Shibagaki N, Shinohara N, Shippen DE, Sorensen I, Sotooka R, Sugimoto N, Sugita M, Sumikawa N, Tanurdzic M, Theissen G, Ulvskov P, Wakazuki S, Weng JK, Willats WW, Wipf D, Wolf PG, Yang L, Zimmer AD, Zhu Q, Mitros T, Hellsten U, Loque D, Otillar R, Salamov A, Schmutz J, Shapiro H, Lindquist E, Lucas S, Rokhsar D, Grigoriev IV ( 2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science, 332, 960-963. |
| 4 | Bao J, Xia H, Zhou J, Liu X, Wang G ( 2013) Efficient implementation of MrBayes on multi-GPU. Molecular Biology and Evolution, 30, 1471-1479. |
| 5 | Barkan A, Small I ( 2014) Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology, 65, 415-442. |
| 6 | Bedbrook JR, Kolodner R, Bogorad L ( 1977) Zea mays chloroplast ribosomal RNA genes are part of a 22,000 base pair inverted repeat. Cell, 11, 739-749. |
| 7 | Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu YL, Kron KA, Rettig JH, Conti E, Palmer JD, Manhart JR, Sytsma KJ, Michaels HJ, Kress WJ, Karol KG, Clark WD, Hedren M, Gaut BS, Jansen RK, Kim KJ, Wimpee CF, Smith JF, Furnier GR, Strauss SH, Xiang QY, Plunkett GM, Soltis PS, Swensen SM, Williams SE, Gadek PA, Quinn CJ, Eguiarte LE, Golenberg E, Learn GH, Graham SW, Barrett SCH, Dayanandan S, Albert VA ( 1993) Phylogenetics of seed plants: An analysis of nucleotide sequences from the plastid gene rbcL. Annals of the Missouri Botanical Garden, 80, 528-580. |
| 8 | Clegg MT, Gaut BS, Learn GH, Morton BR ( 1994) Rates and patterns of chloroplast DNA evolution. Proceedings of the National Academy of Sciences, USA, 91, 6795-6801. |
| 9 | Der JP, Barker MS, Wickett NJ, dePamphilis CW, Wolf PG ( 2011) De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics, 12, 99. |
| 10 | Fuentes P, Armarego-Marriott T, Bock R ( 2018) Plastid transformation and its application in metabolic engineering. Current Opinion in Biotechnology, 49, 10-15. |
| 11 | Gao L, Wang B, Wang ZW, Zhou Y, Su YJ, Wang T ( 2013) Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates. Genome Biology and Evolution, 5, 1403-1407. |
| 12 | Gao L, Yi X, Yang YX, Su YJ, Wang T ( 2009) Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evolutionary Biology, 9, 130. |
| 13 | Gao L, Zhou Y, Wang ZW, Su YJ, Wang T ( 2011) Evolution of the rpoB-psbZ region in fern plastid genomes: Notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biology, 11, 64. |
| 14 | Gitzendanner MA, Soltis PS, Yi TS, Li DZ, Soltis DE ( 2018) Plastome phylogenetics: 30 years of inferences into plant evolution. Advances in Botanical Research, 85, 293-313. |
| 15 | Gott JM, Emeson RB ( 2000) Functions and mechanisms of RNA editing. Annual Review of Genetics, 34, 499-531. |
| 16 | Grewe F, Guo WH, Gubbels EA, Hansen AK, Mower JP ( 2013) Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evolutionary Biology, 13, 8. |
| 17 | Grewe F, Viehoever P, Weisshaar B, Knoop V ( 2009) A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Research, 37, 5093-5104. |
| 18 | Guo W, Zhu A, Fan W, Mower JP ( 2017) Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytologist, 213, 391-403. |
| 19 | Hasebe M, Wolf PG, Pryer KM, Ueda K, Ito M, Sano R, Gastony GJ, Yokoyama J, Manhart JR, Murakami N, Crane EH, Haufler CH, Hauk WD ( 1995) Fern phylogeny based on rbcL nucleotide sequences. American Fern Journal, 85, 134-181. |
| 20 | Hecht J, Grewe F, Knoop V ( 2011) Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biology and Evolution, 3, 344-358. |
| 21 | Hidalgo O, Pellicer J, Christenhusz MJM, Schneider H, Leitch IJ ( 2017) Genomic gigantism in the whisk-fern family (Psilotaceae): Tmesipteris obliqua challenges record holder Paris japonica. Botanical Journal of the Linnean Society, 183, 509-514. |
| 22 | Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, Li YQ, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M ( 1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Molecular and General Genetics, 217, 185-194. |
| 23 | Ichinose M, Sugita M ( 2017) RNA editing and its molecular mechanism in plant organelles. Genes, 8, 5. |
| 24 | Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee S-B, Peery R, McNeal JR, Kuehl JV, Boore JL ( 2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns.Proceedings of the National Academy of Sciences,USA, 104, 19369-19374. |
| 25 | Jansen RK, Ruhlman TA ( 2012) Plastid genomes of seed plants.In: Genomics of Chloroplasts and Mitochondria (eds Bock R, Knoop V), pp.103-126. Springer-Netherlands, Dordrecht. |
| 26 | Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS ( 2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587. |
| 27 | Karol KG, Arumuganathan K, Boore JL, Duffy AM, Everett KD, Hall JD, Hansen SK, Kuehl JV, Mandoli DF, Mishler BD, Olmstead RG, Renzaglia KS, Wolf PG ( 2010) Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evolutionary Biology, 10, 321. |
| 28 | Kim HT, Chung MG, Kim KJ ( 2014) Chloroplast genome evolution in early diverged leptosporangiate ferns. Molecules and Cells, 37, 372-382. |
| 29 | Kim HT, Kim KJ ( 2014) Chloroplast genome differences between Asian and American Equisetum arvense (Equisetaceae) and the origin of the hypervariable trnY-trnE intergenic spacer. PLoS ONE, 9, e103898. |
| 30 | Knox EB ( 2014) The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms.Proceedings of the National Academy of Sciences,USA,111, 11097-11102. |
| 31 | Kolodner R, Tewari KK ( 1979) Inverted repeats in chloroplast DNA from higher plants. Proceedings of the National Academy of Sciences,USA, 76, 41-45. |
| 32 | Kuo LY, Qi X, Ma H, Li FW ( 2018) Order-level fern plastome phylogenomics: New insights from Hymenophyllales. American Journal of Botany, 105, 1545-1555. |
| 33 | Labiak PH, Karol KG ( 2017) Plastome sequences of an ancient fern lineage reveal remarkable changes in gene content and architecture. American Journal of Botany, 104, 1008-1018. |
| 34 | Lenz H, Rüdinger M, Volkmar U, Fischer S, Herres S, Grewe F, Knoop V ( 2010) Introducing the plant RNA editing prediction and analysis computer tool PREPACT and an update on RNA editing site nomenclature. Current Genetics, 56, 189-201. |
| 35 | Li FW, Brouwer P, Carretero-Paulet L, Cheng S, de Vries J, Delaux PM, Eily A, Koppers N, Kuo LY, Li Z, Simenc M, Small I, Wafula E, Angarita S, Barker MS, Bräutigam A, dePamphilis C, Gould S, Hosmani PS, Huang YM, Huettel B, Kato Y, Liu X, Maere S, McDowell R, Mueller LA, Nierop KGJ, Rensing SA, Robison T, Rothfels CJ, Sigel EM, Song Y, Timilsena PR, van de Peer Y, Wang H, Wilhelmsson PKI, Wolf PG, Xu X, Der JP, Schluepmann H, Wong GKS, Pryer KM ( 2018) Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nature Plants, 4, 460-472. |
| 36 | Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, Wang H, van der Bank M, Zhang SD, Wang QF, Wang J, Zhang ZR, Fu CN, Yang J, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Li DZ ( 2019) Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants, 5, 455-456. |
| 37 | Liu L, Shu JP, Wei HJ, Zhang R, Shen H, Yan YH ( 2016) De novo transcriptome analysis of the rare fern Monachosorum maximowiczii (Dennstaedtiaceae) endemic to East Asia. Biodiversity Science, 24, 1325-1334. (in Chinese with English abstract) |
| [ 刘莉, 舒江平, 韦宏金, 张锐, 沈慧, 严岳鸿 ( 2016) 东亚特有珍稀蕨类植物岩穴蕨(碗蕨科)高通量转录组测序及分析. 生物多样性, 24, 1325-1334.] | |
| 38 | Liu Y, Wang B, Cui P, Li L, Xue JY, Yu J, Qiu YL ( 2012) The mitochondrial genome of the lycophyte Huperzia squarrosa: The most archaic form in vascular plants. PLoS ONE, 7, e35168. |
| 39 | Logacheva MD, Krinitsina AA, Belenikin MS, Khafizov K, Konorov EA, Kuptsov SV, Speranskaya AS ( 2017) Comparative analysis of inverted repeats of polypod fern (Polypodiales) plastomes reveals two hypervariable regions. BMC Plant Biology, 17, 255. |
| 40 | Lu JM, Zhang N, Du XY, Wen J, Li DZ ( 2015) Chloroplast phylogenomics resolves key relationships in ferns. Journal of Systematics and Evolution, 53, 448-457. |
| 41 | Ma PF, Zhang YX, Guo ZH, Li DZ ( 2015) Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Scientific Reports, 5, 11608. |
| 42 | Male PJ, Bardon L, Besnard G, Coissac E, Delsuc F, Engel J, Lhuillier E, Scotti-Saintagne C, Tinaut A, Chave J ( 2014) Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Molecular Ecology Resources, 14, 966-975. |
| 43 | Manning JE, Wolstenholme DR, Ryan RS, Hunter JA, Richards OC ( 1971) Circular chloroplast DNA from Euglena gracilis. Proceedings of the National Academy of Sciences,USA, 68, 1169-1173. |
| 44 | Moore MJ, Bell CD, Soltis PS, Soltis DE ( 2007) Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences,USA, 104, 19363-19368. |
| 45 | Mower JP, Ma PF, Grewe F, Taylor A, Michael TP, VanBuren R, Qiu YL ( 2019) Lycophyte plastid genomics: Extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytologist, 222, 1061-1075. |
| 46 | Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ ( 2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268-274. |
| 47 | Nunes GL, Oliveira RRM, Guimaraes JTF, Giulietti AM, Caldeira C, Vasconcelos S, Pires E, Dias M, Watanabe M, Pereira J, Jaffe R, Bandeira C, Carvalho-Filho N, da Silva EF, Rodrigues TM, Dos Santos FMG, Fernandes T, Castilho A, Souza-Filho PWM, Imperatriz-Fonseca V, Siqueira JO, Alves R, Oliveira G ( 2018) Quillworts from the Amazon: A multidisciplinary populational study on Isoetes serracarajensis and Isoetes cangae. PLoS ONE, 13, e0201417. |
| 48 | Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S, Inokuchi H, Ozeki H ( 1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature, 322, 572-574. |
| 49 | Oldenburg DJ, Bendich AJ ( 2015) DNA maintenance in plastids and mitochondria of plants. Frontiers in Plant Science, 6, 883. |
| 50 | Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V ( 2014) Chloroplast RNA editing going extreme: More than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA, 20, 1499-1506. |
| 51 | Palmer JD, Zamir D ( 1982) Chloroplast DNA evolution and phylogenetic relationships in Lycopersicon. Proceedings of the National Academy of Sciences,USA, 79, 5006-5010. |
| 52 | PPG I ( 2016) A community-derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution, 54, 563-603. |
| 53 | Qi X, Kuo LY, Guo C, Li H, Li Z, Qi J, Wang L, Hu Y, Xiang J, Zhang C, Guo J, Huang CH, Ma H ( 2018) A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Molecular Phylogenetics and Evolution, 127, 961-977. |
| 54 | Raubeson LA, Jansen RK ( 1992) Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science, 255, 1697-1699. |
| 55 | Robison TA, Grusz AL, Wolf PG, Mower JP, Fauskee BD, Sosa K, Schuettpelz E ( 2018) Mobile elements shape plastome evolution in ferns. Genome Biology and Evolution, 10, 2558-2571. |
| 56 | Robison TA, Wolf PG ( 2019) ReFernment: An R package for annotating RNA editing in plastid genomes. Applications in Plant Sciences, 7, e01216. |
| 57 | Roper JM, Hansen SK, Wolf PG, Karol KG, Mandoli DF, Everett KDE, Kuehl J, Boore JL ( 2007) The complete plastid genome sequence of Angiopteris evecta (G. Forst.) Hoffm. (Marattiaceae). American Fern Journal, 97, 95-106. |
| 58 | Rothfels CJ, Li FW, Sigel EM, Huiet L, Larsson A, Burge DO, Ruhsam M, Deyholos M, Soltis DE, Stewart CN Jr, Shaw SW, Pokorny L, Chen T, dePamphilis C, DeGironimo L, Chen L, Wei X, Sun X, Korall P, Stevenson DW, Graham SW, Wong GK, Pryer KM ( 2015) The evolutionary history of ferns inferred from 25 low-copy nuclear genes. American Journal of Botany, 102, 1089-1107. |
| 59 | Sager R, Ishida MR ( 1963) Chloroplast DNA in Chlamydomonas. Proceedings of the National Academy of Sciences,USA, 50, 725-730. |
| 60 | Sauquet H, Magallon S ( 2018) Key questions and challenges in angiosperm macroevolution. New Phytologist, 219, 1170-1187. |
| 61 | Schafran PW, Zimmer EA, Taylor WC, Musselman LJ ( 2018) A whole chloroplast genome phylogeny of diploid species of Isoëtes (Isoëtaceae, Lycopodiophyta) in the southeastern United States. Castanea, 83, 224-235. |
| 62 | Sessa EB, Der JP ( 2016) Evolutionary genomics of ferns and lycophytes. Advances in Botanical Research, 78, 215-254. |
| 63 | Shen H, Jin D, Shu JP, Zhou XL, Lei M, Wei R, Shang H, Wei HJ, Zhang R, Liu L, Gu YF, Zhang XC, Yan YH ( 2018) Large scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience, 7, 1-11. |
| 64 | Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M ( 1986) The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. The EMBO Journal, 5, 2043-2049. |
| 65 | Smith DR ( 2009) Unparalleled GC content in the plastid DNA of Selaginella. Plant Molecular Biology, 71, 627-639. |
| 66 | Smith DR ( 2018) Plastid genomes hit the big time. New Phytologist, 219, 491-495. |
| 67 | Song M, Kuo LY, Huiet L, Pryer KM, Rothfels CJ, Li FW ( 2018) A novel chloroplast gene reported for flagellate plants. American Journal of Botany, 105, 117-121. |
| 68 | Sugiura M ( 1992) The chloroplast genome. Plant Molecular Biology, 19, 149-168. |
| 69 | Tsuji S, Ueda K, Nishiyama T, Hasebe M, Yoshikawa S, Konagaya A, Nishiuchi T, Yamaguchi K ( 2007) The chloroplast genome from a lycophyte (microphyllophyte), Selaginella uncinata, has a unique inversion, transpositions and many gene losses. Journal of Plant Research, 120, 281-290. |
| 70 | VanBuren R, Wai CM, Ou S, Pardo J, Bryant D, Jiang N, Mockler TC, Edger P, Michael TP ( 2018) Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla. Nature Communications, 9, 13. |
| 71 | Wakasuki T, Nishikawa A, Yamada K, Sugiura M ( 1998) A complete nucleotide sequence of the plastid genome from a fern, Psilotum nudum. Endocytobiosis and Cell Research, 13(Suppl.), 147. |
| 72 | Wang W, Tanurdzic M, Luo M, Sisneros N, Kim HR, Weng JK, Kudrna D, Mueller C, Arumuganathan K, Carlson J, Chapple C, dePamphilis C, Mandoli D, Tomkins J, Wing RA, Banks JA ( 2005) Construction of a bacterial artificial chromosome library from the spikemoss Selaginella moellendorffii: A new resource for plant comparative genomics. BMC Plant Biology, 5, 10. |
| 73 | Wei R, Yan YH, Harris AJ, Kang JS, Shen H, Xiang QP, Zhang XC ( 2017) Plastid phylogenomics resolve deep relationships among eupolypod II ferns with rapid radiation and rate heterogeneity. Genome Biology and Evolution, 9, 1646-1657. |
| 74 | Wolf PG, Aaron MD, Roper JM ( 2009) Phylogenetic use of inversions in fern chloroplast genomes. American Fern Journal, 99, 132-134. |
| 75 | Wolf PG, Der JP, Duffy AM, Davidson JB, Grusz AL, Pryer KM ( 2011) The evolution of chloroplast genes and genomes in ferns. Plant Molecular Biology, 76, 251-261. |
| 76 | Wolf PG, Karol KG ( 2012) Plastomes of bryophytes, lycophytes and ferns. In: Genomics of Chloroplasts and Mitochondria (eds Bock R, Knoop V), pp.89-102. Springer-Netherlands, Dordrecht. |
| 77 | Wolf PG, Karol KG, Mandoli DF, Kuehl J, Arumuganathan K, Ellis MW, Mishler BD, Kelch DG, Olmstead RG, Boore JL ( 2005) The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene, 350, 117-128. |
| 78 | Wolf PG, Roper JM, Duffy AM ( 2010) The evolution of chloroplast genome structure in ferns. Genome, 53, 731-738. |
| 79 | Wolf PG, Rowe CA, Hasebe M ( 2004) High levels of RNA editing in a vascular plant chloroplast genome: Analysis of transcripts from the fern Adiantum capillus-veneris. Gene, 339, 89-97. |
| 80 | Wolf PG, Rowe CA, Sinclair RB, Hasebe M ( 2003) Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA Research, 10, 59-65. |
| 81 | Wolf PG, Sessa EB, Marchant DB, Li FW, Rothfels CJ, Sigel EM, Gitzendanner MA, Visger CJ, Banks JA, Soltis DE ( 2015) An exploration into fern genome space. Genome Biology and Evolution, 7, 2533-2544. |
| 82 | Xu Z, Xin T, Bartels D, Li Y, Gu W, Yao H, Liu S, Yu H, Pu X, Zhou J, Xu J, Xi C, Lei H, Song J, Chen S ( 2018) Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Molecular Plant, 11, 983-994. |
| 83 | Yang JB, Li DZ, Li HT ( 2014) Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Molecular Ecology Resources, 14, 1024-1031. |
| 84 | Yao G, Jin JJ, Li HT, Yang JB, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, Soltis DE, Soltis PS, Smith SA, Brockington SF, Moore MJ, Yi TS, Li DZ ( 2019) Plastid phylogenomic insights into the evolution of Caryophyllales. Molecular Phylogenetics and Evolution, 134, 74-86. |
| 85 | Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB ( 2018) Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods, 14, 43. |
| 86 | Zhang HR, Xiang QP, Zhang XC ( 2019) The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (Lycophyte). Genome Biology and Evolution, 11, 1258-1274. |
| 87 | Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS ( 2017) Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytologist, 214, 1355-1367. |
| 88 | Zhang YJ, Li DZ ( 2011) Advances in phylogenomics based on complete chloroplast genomes. Plant Diversity and Resources, 33, 365-375. (in Chinese with English abstract) |
| [ 张韵洁, 李德铢 ( 2011) 叶绿体系统发育基因组学的研究进展. 植物分类与资源学报, 33, 365-375.] | |
| 89 | Zhang YJ, Ma PF, Li DZ ( 2011) High-throughput sequencing of six bamboo chloroplast genomes: Phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS ONE, 6, e20596. |
| 90 | Zhong B, Fong R, Collins LJ, McLenachan PA, Penny D ( 2014) Two new fern chloroplasts and decelerated evolution linked to the long generation time in tree ferns. Genome Biology and Evolution, 6, 1166-1173. |
| 91 | Zhu A, Guo W, Gupta S, Fan W, Mower JP ( 2016) Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytologist, 209, 1747-1756. |
| 92 | Zou XH, Ge S ( 2008) Conflicting gene trees and phylogenomics. Journal of Systematics and Evolution, 46, 795-807. (in Chinese with English abstract) |
| [ 邹新慧, 葛颂 ( 2008) 基因树冲突与系统发育基因组学研究. 植物分类学报, 46, 795-807.] |
| [1] | 朱瑞良, 马晓英, 曹畅, 曹子寅. 中国苔藓植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22378-. |
| [2] | 钱宏, 张健, 赵静超. 世界上已知维管植物有多少种? 基于多个全球植物数据库的整合[J]. 生物多样性, 2022, 30(7): 22254-. |
| [3] | 张淑梅, 李微, 李丁男. 辽宁省高等植物多样性编目[J]. 生物多样性, 2022, 30(6): 22038-. |
| [4] | 王婷, 夏增强, 舒江平, 张娇, 王美娜, 陈建兵, 王慷林, 向建英, 严岳鸿. 全基因组复制事件的绝对定年揭示莲座蕨属植物的迟滞演化[J]. 生物多样性, 2021, 29(6): 722-734. |
| [5] | 张梦华, 张宪春. 中国薄叶卷柏复合群的物种划分[J]. 生物多样性, 2021, 29(12): 1607-1619. |
| [6] | 王伟, 刘阳. 植物生命之树重建的现状、问题和对策建议[J]. 生物多样性, 2020, 28(2): 176-188. |
| [7] | 孙晶琦, 陈泉, 李航宇, 常艳芬, 巩合德, 宋亮, 卢华正. 附生蕨类植物的克隆性研究进展[J]. 生物多样性, 2019, 27(11): 1184-1195. |
| [8] | 汪浩, 张锐, 张娇, 沈慧, 戴锡玲, 严岳鸿. 转录组测序揭示翼盖蕨(Didymochlaena trancatula)的全基因组复制历史[J]. 生物多样性, 2019, 27(11): 1221-1227. |
| [9] | 赵国华, 王莹, 商辉, 周喜乐, 王爱华, 李玉峰, 王晖, 刘保东, 严岳鸿. 祖先性状重建法揭示铁线蕨属植物孢子表面纹饰的形态多样性及其演化[J]. 生物多样性, 2019, 27(11): 1228-1235. |
| [10] | 詹臻, 张剑锋, 曹建国, 戴锡玲. 普通针毛蕨颈卵器和卵的发育[J]. 生物多样性, 2019, 27(11): 1236-1244. |
| [11] | 张开梅, 沈羽, 周晓丽, 方炎明. 21世纪以来蕨类植物研究论文的发表情况: 基于Web of Science的数据统计[J]. 生物多样性, 2019, 27(11): 1245-1250. |
| [12] | 姬红利, 詹选怀, 张丽, 彭焱松, 周赛霞, 胡菀. 幕阜山脉石松类和蕨类植物多样性及生物地理学特征[J]. 生物多样性, 2019, 27(11): 1251-1259. |
| [13] | 杜维波, 卢元. 黄土高原石松类和蕨类植物的多样性与地理分布[J]. 生物多样性, 2019, 27(11): 1260-1267. |
| [14] | 董仕勇, 左政裕, 严岳鸿, 向建英. 中国石松类和蕨类植物的红色名录评估[J]. 生物多样性, 2017, 25(7): 765-773. |
| [15] | 常艳芬. 铁角蕨科的多倍化与物种多样性形成[J]. 生物多样性, 2017, 25(6): 621-626. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn