



Biodiv Sci ›› 2023, Vol. 31 ›› Issue (4): 22391. DOI: 10.17520/biods.2022391 cstr: 32101.14.biods.2022391
• Original Papers: Genetic Diversity • Next Articles
Fei Xiong1,2, Hongyan Liu1,2,*( ), Dongdong Zhai1,2, Xinbin Duan3,*(
), Dongdong Zhai1,2, Xinbin Duan3,*( ), Huiwu Tian3, Daqing Chen3
), Huiwu Tian3, Daqing Chen3
Received:2022-07-09
Accepted:2022-11-14
Online:2023-04-20
Published:2023-04-20
Contact:
*E-mail: Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing[J]. Biodiv Sci, 2023, 31(4): 22391.
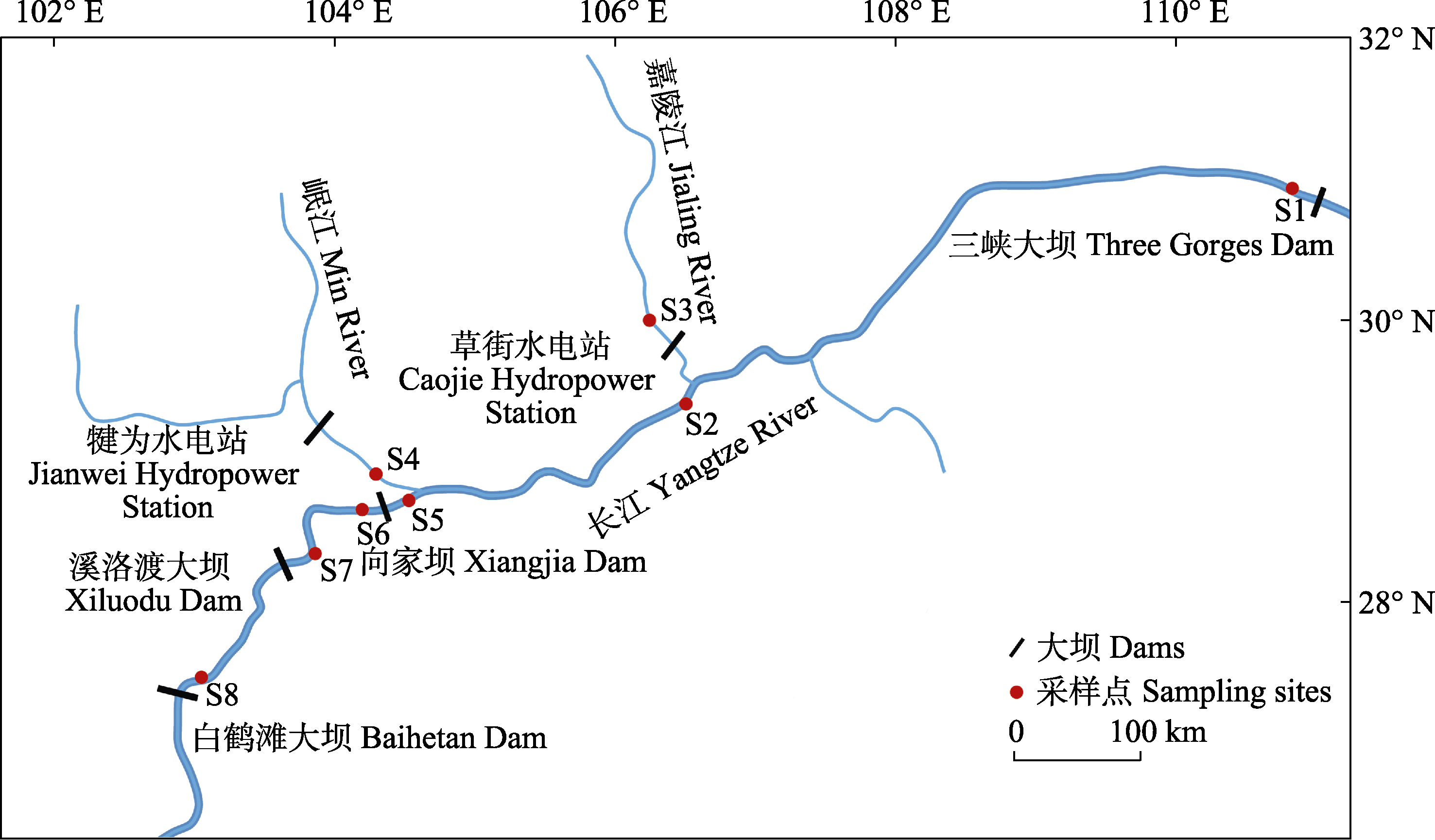
Fig. 1 Sampling sites of Pelteobagrus vachelli in the upper reaches of Yangtze River. S1, Taipingxi (TP); S2, Banan (BN); S3, Hechuan (HC); S4, Minjiangkou (MJ); S5, Yibin (YB); S6, Shaonüping (SN); S7, Huixi (HX); S8, Fengjiaping (FJ).
| 群体 Population | 采样时间 Sampling time | 样本数 No. of sample | 海拔 Elevation (m) | 坡降 Channel slope (m/km) | 生境类型 Habitat type |
|---|---|---|---|---|---|
| 太平溪 TP | 2019.8 | 14 | 110 | 0.12 | 水库 Reservoir |
| 巴南 BN | 2019.8 | 25 | 160 | 0.20 | 回水区 Backwater area |
| 合川 HC | 2019.9 | 4 | 187 | 1.85 | 水库 Reservoir |
| 岷江口 MJ | 2019.10 | 25 | 257 | 2.26 | 河流 River |
| 宜宾 YB | 2019.10 | 6 | 253 | 0.70 | 河流 River |
| 邵女坪 SN | 2019.10 | 12 | 296 | 0.90 | 水库 Reservoir |
| 桧溪 HX | 2019.10 | 25 | 356 | 1.40 | 河流 River |
| 冯家坪 FJ | 2019.10 | 25 | 578 | 1.50 | 河流 River |
Table 1 Sampling information in eight populations of Pelteobagrus vachelli. Site abbreviations are shown in Fig. 1.
| 群体 Population | 采样时间 Sampling time | 样本数 No. of sample | 海拔 Elevation (m) | 坡降 Channel slope (m/km) | 生境类型 Habitat type |
|---|---|---|---|---|---|
| 太平溪 TP | 2019.8 | 14 | 110 | 0.12 | 水库 Reservoir |
| 巴南 BN | 2019.8 | 25 | 160 | 0.20 | 回水区 Backwater area |
| 合川 HC | 2019.9 | 4 | 187 | 1.85 | 水库 Reservoir |
| 岷江口 MJ | 2019.10 | 25 | 257 | 2.26 | 河流 River |
| 宜宾 YB | 2019.10 | 6 | 253 | 0.70 | 河流 River |
| 邵女坪 SN | 2019.10 | 12 | 296 | 0.90 | 水库 Reservoir |
| 桧溪 HX | 2019.10 | 25 | 356 | 1.40 | 河流 River |
| 冯家坪 FJ | 2019.10 | 25 | 578 | 1.50 | 河流 River |
| 群体 Population | 样本数 No. of sample | Clean data (Gb) | Q30 (%) | 测序深度Sequencing depth | 序列比对率 Sequence mapped (%) | 基因组覆盖度Coverage of genomic reference (%) | SNP数目 Number of SNP | 核苷酸多样性指数Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| 太平溪 TP | 14 | 145.22 | 93.11 | 9.54 | 54.31 | 72.92 | 4,172,876 | 2.44 × 10?3 |
| 巴南 BN | 25 | 236.41 | 92.82 | 8.71 | 53.43 | 71.68 | 3,543,176 | 2.81 × 10?3 |
| 合川 HC | 4 | 35.77 | 92.81 | 8.38 | 49.94 | 66.28 | 534,504 | 8.59 × 10?4 |
| 岷江口 MJ | 25 | 217.32 | 92.73 | 8.25 | 50.68 | 66.39 | 419,733 | 5.60 × 10?4 |
| 宜宾 YB | 6 | 50.16 | 91.87 | 7.44 | 48.50 | 67.36 | 563,968 | 6.76 × 10?4 |
| 邵女坪 SN | 12 | 128.29 | 92.79 | 9.37 | 49.09 | 69.64 | 457,603 | 5.75 × 10?4 |
| 桧溪 HX | 25 | 258.76 | 92.79 | 9.20 | 49.22 | 68.87 | 597,465 | 6.60 × 10?4 |
| 冯家坪 FJ | 25 | 273.27 | 92.78 | 9.57 | 49.06 | 69.73 | 441,092 | 5.84 × 10?4 |
| 总体 Total | 136 | 1,345.21 | 92.78 | 8.95 | 50.73 | 69.43 | 7,341,959 | 1.19 × 10?3 |
Table 2 The genome re-sequencing data and genetic diversity parameters in eight populations of Pelteobagrus vachelli
| 群体 Population | 样本数 No. of sample | Clean data (Gb) | Q30 (%) | 测序深度Sequencing depth | 序列比对率 Sequence mapped (%) | 基因组覆盖度Coverage of genomic reference (%) | SNP数目 Number of SNP | 核苷酸多样性指数Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| 太平溪 TP | 14 | 145.22 | 93.11 | 9.54 | 54.31 | 72.92 | 4,172,876 | 2.44 × 10?3 |
| 巴南 BN | 25 | 236.41 | 92.82 | 8.71 | 53.43 | 71.68 | 3,543,176 | 2.81 × 10?3 |
| 合川 HC | 4 | 35.77 | 92.81 | 8.38 | 49.94 | 66.28 | 534,504 | 8.59 × 10?4 |
| 岷江口 MJ | 25 | 217.32 | 92.73 | 8.25 | 50.68 | 66.39 | 419,733 | 5.60 × 10?4 |
| 宜宾 YB | 6 | 50.16 | 91.87 | 7.44 | 48.50 | 67.36 | 563,968 | 6.76 × 10?4 |
| 邵女坪 SN | 12 | 128.29 | 92.79 | 9.37 | 49.09 | 69.64 | 457,603 | 5.75 × 10?4 |
| 桧溪 HX | 25 | 258.76 | 92.79 | 9.20 | 49.22 | 68.87 | 597,465 | 6.60 × 10?4 |
| 冯家坪 FJ | 25 | 273.27 | 92.78 | 9.57 | 49.06 | 69.73 | 441,092 | 5.84 × 10?4 |
| 总体 Total | 136 | 1,345.21 | 92.78 | 8.95 | 50.73 | 69.43 | 7,341,959 | 1.19 × 10?3 |
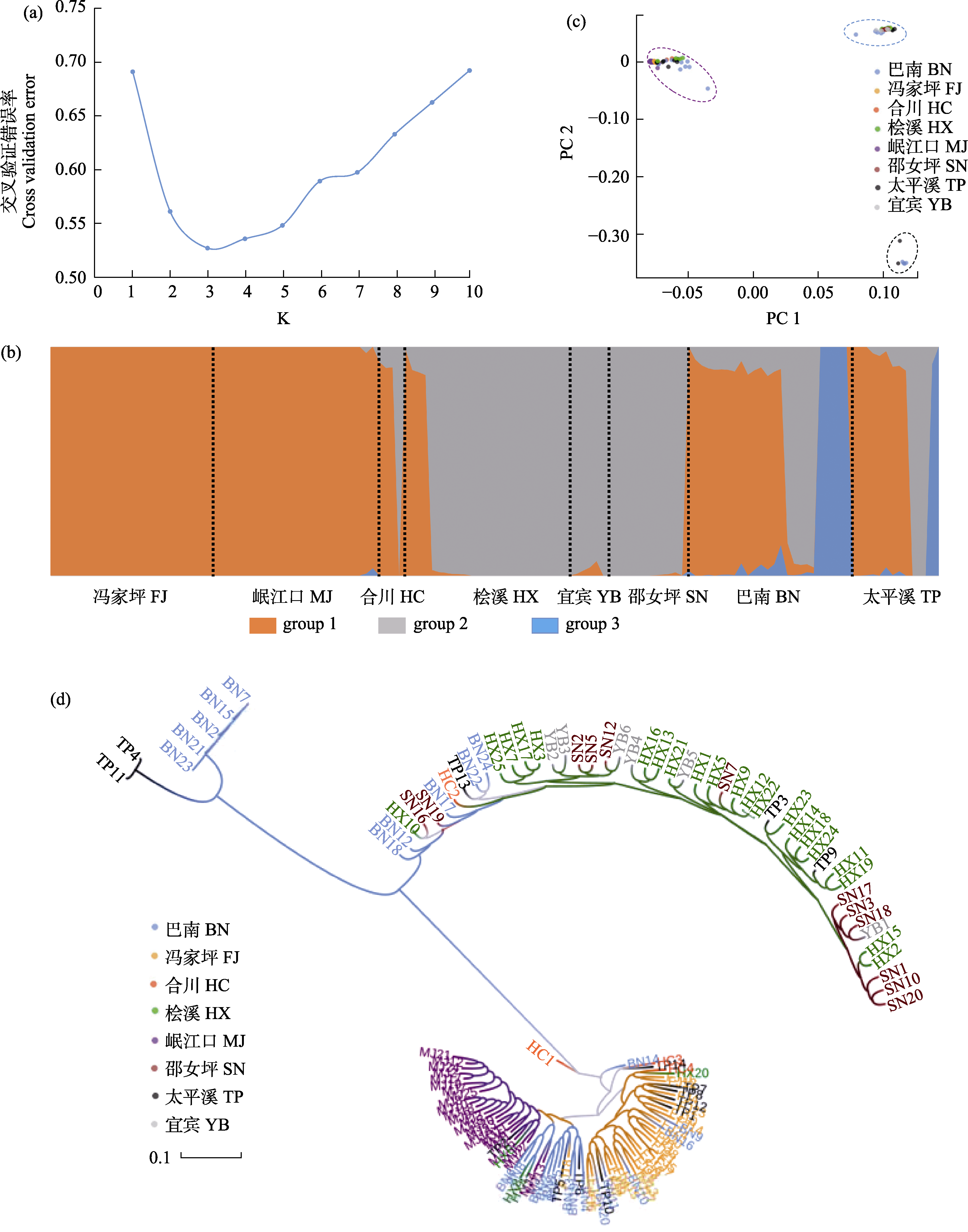
Fig. 2 Genetic structure based on SNP markers in eight populations of Pelteobagrus vaclerii. (a) The optimal number of clusters determined by cross-validated error rate valley. (b) Population structure (K = 3) of all individuals. (c) PCA plot of every individual. (d) ML phylogenetic tree. Site abbreviations are shown in Fig. 1.
| 群体 Population | 太平溪 TP | 巴南 BN | 合川 HC | 岷江口 MJ | 宜宾 YB | 邵女坪 SN | 桧溪 HX |
|---|---|---|---|---|---|---|---|
| 巴南 BN | 0.050 | ||||||
| 合川 HC | ?0.109 | ?0.039 | |||||
| 岷江口 MJ | 0.112 | 0.143 | 0.075 | ||||
| 宜宾 YB | ?0.020 | 0.043 | 0.152 | 0.233 | |||
| 邵女坪 SN | 0.068 | 0.112 | 0.192 | 0.235 | 0.000 | ||
| 桧溪 HX | 0.126 | 0.154 | 0.079 | 0.179 | ?0.008 | 0.002 | |
| 冯家坪 FJ | 0.125 | 0.152 | 0.092 | 0.016 | 0.261 | 0.257 | 0.193 |
Table 3 Genetic differentiation between the pairwise populations of Pelteobagrus vaclerii in the upper Yangtze River
| 群体 Population | 太平溪 TP | 巴南 BN | 合川 HC | 岷江口 MJ | 宜宾 YB | 邵女坪 SN | 桧溪 HX |
|---|---|---|---|---|---|---|---|
| 巴南 BN | 0.050 | ||||||
| 合川 HC | ?0.109 | ?0.039 | |||||
| 岷江口 MJ | 0.112 | 0.143 | 0.075 | ||||
| 宜宾 YB | ?0.020 | 0.043 | 0.152 | 0.233 | |||
| 邵女坪 SN | 0.068 | 0.112 | 0.192 | 0.235 | 0.000 | ||
| 桧溪 HX | 0.126 | 0.154 | 0.079 | 0.179 | ?0.008 | 0.002 | |
| 冯家坪 FJ | 0.125 | 0.152 | 0.092 | 0.016 | 0.261 | 0.257 | 0.193 |
| [1] |
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655-1664.
DOI PMID |
| [2] | Chen DQ, Liu SP, Duan XB, Xiong F (2002) A preliminary study of the fisheries biology of main commercial fishes in the middle and upper reaches of the Yangtze River. Acta Hydrobiologica Sinica, 26, 618-622. (in Chinese with English abstract) |
| [ 陈大庆, 刘绍平, 段辛斌, 熊飞 (2002) 长江中上游主要经济鱼类的渔业生物学特征. 水生生物学报, 26, 618-622.] | |
| [3] |
Cheng F, Li W, Castello L, Murphy BR, Xie SG (2015) Potential effects of dam cascade on fish: Lessons from the Yangtze River. Reviews in Fish Biology and Fisheries, 25, 569-585.
DOI URL |
| [4] |
Cheng F, Li W, Wu QJ, Hallerman E, Xie SG (2013) Microsatellite DNA variation among samples of bronze gudgeon, Coreius heterodon, in the mainstem of the Yangtze River, China. Ichthyological Research, 60, 165-171.
DOI URL |
| [5] |
Cook DR, Sullivan SMP (2018) Associations between riffle development and aquatic biota following low head dam removal. Environmental Monitoring and Assessment, 190, 339.
DOI |
| [6] |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156-2158.
DOI PMID |
| [7] |
Deiner K, Garza JC, Coey R, Girman DJ (2007) Population structure and genetic diversity of trout (Oncorhynchus mykiss) above and below natural and man-made barriers in the Russian River, California. Conservation Genetics, 8, 437-454.
DOI URL |
| [8] | Ding RH (1994) The Fishes of Sichuan. Sichuan Science & Technology Press, Chengdu. (in Chinese) |
| [ 丁瑞华 (1994) 四川鱼类志. 四川科学技术出版社, 成都.] | |
| [9] |
Dixon P (2003) Vegan, a package of R functions for community ecology. Journal of Vegetation Science, 14, 927-930.
DOI URL |
| [10] | Gong GR, Dan C, Xiao SJ, Guo WJ, Huang PP, Xiong Y, Wu JJ, He Y, Zhang JC, Li XH, Chen NS, Gui JF, Mei J (2018) Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. GigaScience, 7, giy120. |
| [11] |
Kang JL, Ma XH, He SP (2017) Population genetics analysis of the Nujiang catfish Creteuchiloglanis macropterus through a genome-wide single nucleotide polymorphisms resource generated by RAD-seq. Scientific Reports, 7, 2813.
DOI |
| [12] |
Kanno Y, Vokoun JC, Letcher BH (2011) Fine-scale population structure and riverscape genetics of brook trout (Salvelinus fontinalis) distributed continuously along headwater channel networks. Molecular Ecology, 20, 3711-3729.
DOI PMID |
| [13] |
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nature Methods, 9, 357-359.
DOI PMID |
| [14] | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Proc GPD (2009) The sequence alignment/map format and SAMtools. Bioinfor- matics, 25, 2078-2079. |
| [15] |
Liu D, Li X, Song Z (2020) No decline of genetic diversity in elongate loach (Leptobotia elongata) with a tendency to form population structure in the upper Yangtze River. Global Ecology and Conservation, 23, e01072.
DOI URL |
| [16] | Liu HY, Xiong F, Duan XB, Tian HW, Liu SP, Chen DQ (2017) Low population differentiation revealed in the highly threatened elongate loach (Leptobotia elongata Bleeker), a species endemic to the fragmented upper reaches of the Yangtze River. Biochemical Systematics & Ecology, 70, 22-28. |
| [17] | Lü H, Tian HW, Duan XB, Chen DQ, Shen SY, Liu SP (2018) The larval resources of fishes spawning drifting eggs in the lower reaches of the Minjiang River. Resources and Environment in the Yangtze Basin, 27(1), 88-96. (in Chinese with English abstract) |
| [ 吕浩, 田辉伍, 段辛斌, 陈大庆, 申绍祎, 刘绍平 (2018) 岷江下游干流段鱼类资源现状及其多样性分析. 长江流域资源与环境, 27(1), 88-96.] | |
| [18] | McKenna A, Hanna M, Banks E, Sivachenk AO, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Resources, 20, 1297-1303. |
| [19] |
Nakajima N, Hirota SK, Matsuo A, Suyama Y, Nakamura F (2020) Genetic structure and population demography of white-spotted charr in the upstream watershed of a large dam. Water, 12, 2406.
DOI URL |
| [20] |
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38, 904-909.
DOI PMID |
| [21] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Bakker PIWD, Daly MJ, Sham PC (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81, 559-575.
DOI URL |
| [22] | Song J, Chang LJ, Wang D (2016) Multi-locus genomic analysis reveals the genetic diversity and population structure of the rock carp (Procypris rabaudi) in the upper Yangtze River. Biochemical Systematics & Ecology, 66, 86-93. |
| [23] |
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312-1313.
DOI PMID |
| [24] |
Vera-Escalona I, Habit E, Ruzzante DE (2015) Echoes of a distant time: Effects of historical processes on contemporary genetic patterns in Galaxias platei in Patagonia. Molecular Ecology, 24, 4112-4128.
DOI PMID |
| [25] |
Wang ZW, Wu QJ, Zhou JF, Ye YZ (2004) Geographic distribution of Pelteobagrus fulvidraco and Pelteobagrus vachelli in the Yangtze River based on mitochondrial DNA markers. Biochemical Genetics, 42, 391-400.
DOI URL |
| [26] | Xiong F, Liu HY, Duan XB, Liu SP, Chen DQ (2015) Present status of fishery resources in Yibin section of the Upper Yangtze River. Journal of Southwest University (Natural Science Edition), 37(11), 43-50. (in Chinese with English abstract) |
| [ 熊飞, 刘红艳, 段辛斌, 刘绍平, 陈大庆 (2015) 长江上游宜宾江段渔业资源现状研究. 西南大学学报(自然科学版), 37(11), 43-50.] | |
| [27] |
Xu SY, Song N, Zhao LL, Cai SS, Han ZQ, Gao TX (2017) Genomic evidence for local adaptation in the ovoviviparous marine fish Sebastiscus marmoratus with a background of population homogeneity. Scientific Reports, 7, 1562.
DOI |
| [28] | Yang JY (1994) The reproductive biology of Pelteobagrus vachelli in the Jialing River. Journal of Southwest China Normal University (Natural Science), 19, 639-645. (in Chinese with English abstract) |
| [ 杨家云 (1994) 嘉陵江瓦氏黄颡鱼的繁殖生物学. 西南师范大学学报(自然科学版), 19, 639-645.] | |
| [29] | Yao WZ (2018) Habitat and restoration of fluvial fish which producing sticky eggs: A case study in the upper reaches of the Yangtze River. In:Proceedings of the Second International Symposium on Modern Marine Ranching and the Fisheries Resources and Environment Committee of the Chinese Aquatic Society, pp. 100. China Aquatic Society Marine Ranching Research Association, Dalian. (in Chinese with English abstract) |
| [ 姚维志 (2018) 河流产粘性卵鱼类生境及修复——以长江上游为例. 见: 第二届现代化海洋牧场国际学术研讨会、中国水产学会渔业资源与环境专业委员会2018年学术年会论文集, pp.100. 中国水产学会海洋牧场研究会, 大连.] | |
| [30] |
Zhai DD, Zhang Z, Zhang FT, Liu HZ, Cao WX, Gao X (2019) Genetic diversity and population structure of a cyprinid fish (Ancherythroculter nigrocauda) in a highly fragmented river. Journal of Applied Ichthyology, 35, 701-708.
DOI URL |
| [31] | Zhang LW, Tian HW, Chen DQ, Liu SP, Duan XB, Wang DQ (2020) Genetic diversity and structure of Glyptothorax sinensis in upper Yangtze River. Freshwater Fisheries, 50, 3-11. (in Chinese with English abstract) |
| [ 张力文, 田辉伍, 陈大庆, 刘绍平, 段辛斌, 汪登强 (2020) 长江上游中华纹胸鮡遗传多样性及遗传结构研究. 淡水渔业, 50, 3-11.] |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [10] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [11] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [12] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [13] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| [14] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| [15] | Jun Sun, Yuyao Song, Yifeng Shi, Jian Zhai, Wenzhuo Yan. Progress of marine biodiversity studies in China seas in the past decade [J]. Biodiv Sci, 2022, 30(10): 22526-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()