



Biodiv Sci ›› 2022, Vol. 30 ›› Issue (2): 21258. DOI: 10.17520/biods.2021258 cstr: 32101.14.biods.2021258
• Original Papers: Genetic Diversity • Previous Articles Next Articles
Jia Song1, Mingyang Zhi1, Qiang Chen2, Yueying Li1, Longkun Wu3, Baoxuan Nong4, Danting Li4, Hongbo Pang1,*( ), Xiaoming Zheng5,*(
), Xiaoming Zheng5,*( )
)
Received:2021-06-29
Accepted:2021-10-14
Online:2022-02-20
Published:2022-02-28
Contact:
Hongbo Pang,Xiaoming Zheng
Jia Song, Mingyang Zhi, Qiang Chen, Yueying Li, Longkun Wu, Baoxuan Nong, Danting Li, Hongbo Pang, Xiaoming Zheng. Nucleotide diversity and adaptation of CTB4a gene related to cold tolerance in rice[J]. Biodiv Sci, 2022, 30(2): 21258.

Fig. 1 Nucleotide diversity in the CTB4a coding region. A, Nucleotide diversity in the coding region of CTB4a gene in 133 cultivated rice samples; B, Amino acid sequence and domains encoded by CTB4a gene, the gray part shows the LRR-RLK domain; C, Nucleotide diversity in the Coding Region of 35 wild rice. Amino acid variation sites in gray. In A and C, Amino acid variation sites (SNP and InDel) and average yellow leaf rate (LYR) values of indica rice population in two groups were shown in gray. And the first column of A and C shows the names of 33 haplotypes of CTB4a gene and 33 wild rice, respectively. cvs, Cultivars; LYR, Yellow leaf rate; I, O. sativa ssp. indica; J, O. sativa ssp. japonica.
| 元件名称 Name | 元件功能 Function | 元件存在类型 Existence type | 元件数量 Number |
|---|---|---|---|
| LTR | 低温诱导 Low-temperature induction | 耐寒/不耐寒 Cold tolerance/intolerance | 1 |
| ABRE | 脱落酸诱导 Abscisic acid induction | 3 | |
| AE-box | 光诱导 Light induction | 10 | |
| G-box | |||
| Box 4 | |||
| Box II | |||
| GATA-motif | |||
| TCCC-motif | |||
| TCT-motif | |||
| ACA-motif | 耐寒 Cold tolerance | 1 | |
| MBS | 干旱诱导 Drought induction | 不耐寒 Cold intolerance | 1 |
Table 1 Cis-acting elements analysis of CTB4a gene promoter
| 元件名称 Name | 元件功能 Function | 元件存在类型 Existence type | 元件数量 Number |
|---|---|---|---|
| LTR | 低温诱导 Low-temperature induction | 耐寒/不耐寒 Cold tolerance/intolerance | 1 |
| ABRE | 脱落酸诱导 Abscisic acid induction | 3 | |
| AE-box | 光诱导 Light induction | 10 | |
| G-box | |||
| Box 4 | |||
| Box II | |||
| GATA-motif | |||
| TCCC-motif | |||
| TCT-motif | |||
| ACA-motif | 耐寒 Cold tolerance | 1 | |
| MBS | 干旱诱导 Drought induction | 不耐寒 Cold intolerance | 1 |
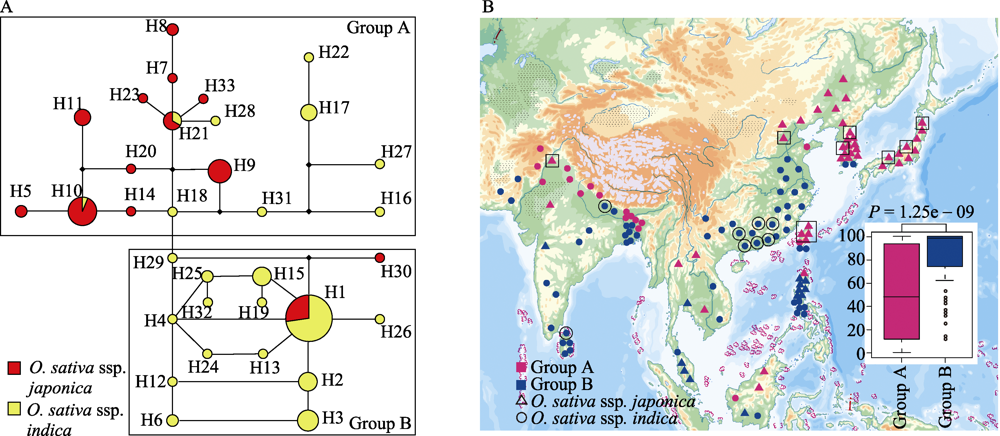
Fig. 2 Network of haplotypes of CTB4a coding region (A) and geographic distribution map in cultivated rice (B). A, The network of all haplotypes for CTB4a, O. sativa ssp. japonica in red and O. sativa ssp. indica in yellow; B, Geographic distribution map of cultivated rice in group A and group B. Group A in pink and Group B in blue; O. sativa ssp. japonica with triangle and O. sativa ssp. indica with circle; H9 haplotype in square; H3 haplotype in circle.
| [1] |
Andaya VC, Mackill DJ (2003a) Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. Journal of Experimental Botany, 54, 2579-2585.
DOI URL |
| [2] |
Andaya VC, Mackill DJ (2003b) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theoretical and Applied Genetics, 106, 1084-1090.
DOI URL |
| [3] |
Chang TT (1976) The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica, 25, 425-441.
DOI URL |
| [4] |
Chory J, Wu DY (2001) Weaving the complex web of signal transduction. Plant Physiology, 125, 77-80.
PMID |
| [5] |
Conley TR, Park SC, Kwon HB, Peng HP, Shih MC (1994) Characterization of cis-acting elements in light regulation of the nuclear gene encoding the A subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Molecular and Cellular Biology, 14, 2525-2533.
DOI PMID |
| [6] |
Cruz RPD, Sperotto RA, Cargnelutti D, Adamski JM, FreitasTerra T, Fett JP (2013) Avoiding damage and achieving cold tolerance in rice plants. Food and Energy Security, 2, 96-119.
DOI URL |
| [7] |
Ding YL, Shi YT, Yang SH (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytologist, 222, 1690-1704.
DOI URL |
| [8] |
Guo HF, Zeng YW, Li JL, Ma XQ, Zhang ZY, Lou QJ, Li J, Gu YS, Zhang HL, Li JJ, Li ZC (2020) Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnology Journal, 18, 2491-2503.
DOI URL |
| [9] |
Guo XY, Liu DF, Chong K (2018) Cold signaling in plants: Insights into mechanisms and regulation. Journal of Integrative Plant Biology, 60, 745-756.
DOI URL |
| [10] |
Hedrick PW (2013) Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology, 22, 4606-4618.
DOI PMID |
| [11] |
Huang CL, Hung CY, Chiang YC, Hwang CC, Hsu TW, Huang CC, Hung KH, Tsai KC, Wang KH, Osada N, Schaal BA, Chiang TY (2012) Footprints of natural and artificial selection for photoperiod pathway genes in Oryza. The Plant Journal, 70, 769-782.
DOI URL |
| [12] |
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology, 35, 25-34.
PMID |
| [13] |
Kim SI, Andaya VC, Tai TH (2011) Cold sensitivity in rice (Oryza sativa L.) is strongly correlated with a naturally occurring I99V mutation in the multifunctional glutathione transferase isoenzyme GSTZ2. The Biochemical Journal, 435, 373-380.
DOI URL |
| [14] |
Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends in Genetics, 23, 578-587.
DOI URL |
| [15] |
Li JL, Zeng YW, Pan YH, Zhou L, Zhang ZY, Guo HF, Lou QJ, Shui GH, Huang HG, Tian H, Guo YM, Yuan PR, Yang H, Pan GJ, Wang RY, Zhang HL, Yang SH, Guo Y, Ge S, Li JJ, Li ZC (2021) Stepwise selection of natural variations at CTB2 and CTB4a improves cold adaptation during domestication of japonica rice. New Phytologist, 231, 1056-1072.
DOI URL |
| [16] |
Liu CT, Ou SJ, Mao BG, Tang JY, Wang W, Wang HR, Cao SY, Schläppi MR, Zhao BR, Xiao GY, Wang XP, Chu CC (2018) Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nature Communications, 9, 3302.
DOI URL |
| [17] |
Liu FX, Sun CQ, Tan LB, Fu YC, Li DJ, Wang XK (2003) Identification and mapping of quantitative trait loci controlling cold-tolerance of Chinese common wild rice (O. rufipogon Griff.) at booting to flowering stages. Chinese Science Bulletin, 48, 2068-2071.
DOI URL |
| [18] |
Lv Y, Hussain MA, Luo D, Tang N (2019) Current understanding of genetic and molecular basis of cold tolerance in rice. Molecular Breeding, 39, 159.
DOI URL |
| [19] |
Ma Y, Dai XY, Xu YY, Luo W, Zheng XM, Zeng DL, Pan YJ, Lin XL, Liu HH, Zhang DJ, Xiao J, Guo XY, Xu SJ, Niu YD, Jin JB, Zhang H, Xu X, Li LG, Wang W, Qian Q, Ge S, Chong K (2015) COLD1 confers chilling tolerance in rice. Cell, 160, 1209-1221.
DOI URL |
| [20] | Mao DH, Xin YY, Tan YJ, Hu XJ, Bai JJ, Liu ZY, Yu YL, Li LY, Peng C, Fan T, Zhu YX, Guo YL, Wang SH, Lu DP, Xing YZ, Yuan LP, Chen CY (2019) Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proceedings of the National Academy of Sciences, USA, 116, 3494-3501. |
| [21] |
Mazzella MA, Bertero D, Casal JJ (2000) Temperature- dependent internode elongation in vegetative plants of Arabidopsis thaliana lacking phytochrome B and cryptochrome 1. Planta, 210, 497-501.
PMID |
| [22] | Oka HI (1958) Intervarietal variation and classification of cultivated rice. Indian Journal of Genetics Plant Breeding, 18, 79-89. |
| [23] |
Penfield S (2008) Temperature perception and signal transduction in plants. New Phytologist, 179, 615-628.
DOI PMID |
| [24] |
Roy S, Choudhury SR, Singh SK, Das KP (2012) Functional analysis of light-regulated promoter region of AtPolλ gene. Planta, 235, 411-432.
DOI URL |
| [25] |
Saito K, Hayano-Saito Y, Kuroki M, Sato Y (2010) Map-based cloning of the rice cold tolerance gene Ctb1. Plant Science, 179, 97-102.
DOI URL |
| [26] |
Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Current Opinion in Plant Biology, 3, 138-142.
PMID |
| [27] |
Shirasawa S, Endo T, Nakagomi K, Yamaguchi M, Nishio T (2012) Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar, ‘Lijiangxin tuanheigu’, in rice, Oryza sativa L. Theoretical and Applied Genetics, 124, 937-946.
DOI PMID |
| [28] |
Sthapit BR, Witcombe JR (1998) Inheritance of tolerance to chilling stress in rice during germination and plumule greening. Crop Science, 38, 660-665.
DOI URL |
| [29] |
Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R (1998) Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA, 4, 1549-1568.
PMID |
| [30] |
Xiao N, Gao Y, Qian HJ, Gao Q, Wu YY, Zhang DP, Zhang XX, Yu L, Li YH, Pan CH, Liu GQ, Zhou CH, Jiang M, Huang NS, Dai ZY, Liang CZ, Chen Z, Chen JM, Li AH (2018) Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiology, 177, 1108-1123.
DOI URL |
| [31] |
Xie GS, Kato H, Imai R (2012) Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. The Biochemical Journal, 443, 95-102.
DOI URL |
| [32] |
Zhang ZY, Li JJ, Pan YH, Li JL, Zhou L, Shi HL, Zeng YW, Guo HF, Yang SM, Zheng WW, Yu JP, Sun XM, Li GL, Ding YL, Ma L, Shen SQ, Dai LY, Zhang HL, Yang SH, Guo Y, Li ZC (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nature Communications, 8, 14788.
DOI URL |
| [33] |
Zhao J, Wang SS, Qin JJ, Sun CQ, Liu FX (2020) The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnology Journal, 18, 756-769.
DOI PMID |
| [34] |
Zhao JL, Zhang SH, Dong JF, Yang TF, Mao XX, Liu Q, Wang XF, Liu B (2017) A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnology Journal, 15, 1141-1148.
DOI URL |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn