



Biodiv Sci ›› 2018, Vol. 26 ›› Issue (11): 1168-1179. DOI: 10.17520/biods.2018223 cstr: 32101.14.biods.2018223
• Original Papers: Plant Diversity • Previous Articles Next Articles
Xingtong Wu1, Lu Chen1, Minqiu Wang1, Yuan Zhang1, Xueying Lin1, Xinyu Li1, Hong Zhou2, Yafeng Wen1,*( )
)
Received:2018-08-13
Accepted:2018-11-28
Online:2018-11-20
Published:2019-01-08
Contact:
Wen Yafeng
About author:# Co-first authors
Xingtong Wu, Lu Chen, Minqiu Wang, Yuan Zhang, Xueying Lin, Xinyu Li, Hong Zhou, Yafeng Wen. Population structure and genetic divergence in Firmiana danxiaensis[J]. Biodiv Sci, 2018, 26(11): 1168-1179.
| 组群 Group | 群体 Population | 编号 Codes | 纬度 Latitude (N) | 经度 Longitude (E) | 数量 Number |
|---|---|---|---|---|---|
| 丹霞山 Danxiashan (DXS) | 锦湖 Jinhu | JH | 25°2°20.24? | 113°44°52.80? | 23 |
| 老苗圃 Laomiaopu | LMP | 25°2°22.24? | 113°44°59.51? | 21 | |
| 韶石山 Shaoshishan | SS | 24°56°43.33? | 113°45°17.60? | 15 | |
| 阳元山 Yangyuanshan | YYS | 25°2°46.08? | 113°44°7.32? | 30 | |
| 长老峰 Zhanglaofeng | ZLF | 25°1°34.70? | 113°44°19.87? | 29 | |
| 南雄 Nanxiong (NX) | 鸳鸯湖 Yuanyanghu | QWT | 25°6°58.53? | 114°11°30.70? | 27 |
| 水库1 Shuiku1 | SK1 | 25°6°58.63? | 114°12°25.62? | 23 | |
| 水库2 Shuiku2 | SK2 | 25°7°12.58? | 114°12°30.57? | 21 | |
| 柴岭 Chailing | WT | 25°7°17.53? | 114°12°16.21? | 24 |
Table 1 Locality information and sample size of the nine Firmiana danxiaensis populations
| 组群 Group | 群体 Population | 编号 Codes | 纬度 Latitude (N) | 经度 Longitude (E) | 数量 Number |
|---|---|---|---|---|---|
| 丹霞山 Danxiashan (DXS) | 锦湖 Jinhu | JH | 25°2°20.24? | 113°44°52.80? | 23 |
| 老苗圃 Laomiaopu | LMP | 25°2°22.24? | 113°44°59.51? | 21 | |
| 韶石山 Shaoshishan | SS | 24°56°43.33? | 113°45°17.60? | 15 | |
| 阳元山 Yangyuanshan | YYS | 25°2°46.08? | 113°44°7.32? | 30 | |
| 长老峰 Zhanglaofeng | ZLF | 25°1°34.70? | 113°44°19.87? | 29 | |
| 南雄 Nanxiong (NX) | 鸳鸯湖 Yuanyanghu | QWT | 25°6°58.53? | 114°11°30.70? | 27 |
| 水库1 Shuiku1 | SK1 | 25°6°58.63? | 114°12°25.62? | 23 | |
| 水库2 Shuiku2 | SK2 | 25°7°12.58? | 114°12°30.57? | 21 | |
| 柴岭 Chailing | WT | 25°7°17.53? | 114°12°16.21? | 24 |
| 位点 Locus | 等位基 因数 Number of allele (Na) | 总遗传多样性 Total genetic diversity for the species (Ht) | 群体内遗传多样性 Genetic diversity within populations (Hs) | 遗传分化系数 Interpersonal inbreeding coefficient (FST) | 标准遗传分 化系数 Gene differentiation factor (GST) | 多态性信息含量 Polymorphism information content (PIC) | 登录号 Accession number |
|---|---|---|---|---|---|---|---|
| unigene_21904 | 9 | 0.651 | 0.590 | 0.102 | 0.094 | 0.577 | MH053446 |
| unigene_9882 | 9 | 0.670 | 0.546 | 0.217 | 0.185 | 0.608 | MH053447 |
| unigene_33644 | 7 | 0.726 | 0.618 | 0.171 | 0.148 | 0.680 | MH053448 |
| unigene_15157 | 8 | 0.800 | 0.722 | 0.094 | 0.098 | 0.765 | MH053449 |
| unigene_23055 | 9 | 0.562 | 0.500 | 0.136 | 0.111 | 0.486 | MH053450 |
| unigene_60653 | 5 | 0.692 | 0.598 | 0.153 | 0.137 | 0.639 | MH053451 |
| unigene_30066 | 6 | 0.588 | 0.554 | 0.064 | 0.057 | 0.498 | MH053452 |
| unigene_37667 | 3 | 0.402 | 0.320 | 0.223 | 0.205 | 0.331 | MH053453 |
| unigene_25213 | 4 | 0.578 | 0.510 | 0.135 | 0.118 | 0.515 | MH053454 |
| unigene_36790 | 13 | 0.745 | 0.582 | 0.240 | 0.219 | 0.717 | MH053455 |
| unigene_5197 | 9 | 0.638 | 0.514 | 0.219 | 0.194 | 0.611 | MH053456 |
| unigene_48018 | 5 | 0.525 | 0.506 | 0.038 | 0.035 | 0.419 | MH053457 |
| unigene_31908 | 15 | 0.700 | 0.640 | 0.080 | 0.085 | 0.659 | MH053458 |
| unigene_25861 | 4 | 0.526 | 0.523 | 0.007 | 0.006 | 0.418 | MH053459 |
| unigene_42278 | 7 | 0.745 | 0.632 | 0.165 | 0.152 | 0.704 | MH053460 |
| unigene_35652 | 10 | 0.733 | 0.597 | 0.205 | 0.186 | 0.711 | MH053461 |
| Fir_SSR25d-F | 5 | 0.549 | 0.478 | 0.126 | 0.130 | 0.475 | KF048045 |
| Fir_SSR81d-F | 3 | 0.527 | 0.398 | 0.285 | 0.244 | 0.422 | KF048051 |
| 均值 Mean | 7.278 | 0.631 | 0.546 | 0.150 | 0.135 | 0.569 |
Table 2 Genetic diversity of 18 loci for Firmiana danxiaensis
| 位点 Locus | 等位基 因数 Number of allele (Na) | 总遗传多样性 Total genetic diversity for the species (Ht) | 群体内遗传多样性 Genetic diversity within populations (Hs) | 遗传分化系数 Interpersonal inbreeding coefficient (FST) | 标准遗传分 化系数 Gene differentiation factor (GST) | 多态性信息含量 Polymorphism information content (PIC) | 登录号 Accession number |
|---|---|---|---|---|---|---|---|
| unigene_21904 | 9 | 0.651 | 0.590 | 0.102 | 0.094 | 0.577 | MH053446 |
| unigene_9882 | 9 | 0.670 | 0.546 | 0.217 | 0.185 | 0.608 | MH053447 |
| unigene_33644 | 7 | 0.726 | 0.618 | 0.171 | 0.148 | 0.680 | MH053448 |
| unigene_15157 | 8 | 0.800 | 0.722 | 0.094 | 0.098 | 0.765 | MH053449 |
| unigene_23055 | 9 | 0.562 | 0.500 | 0.136 | 0.111 | 0.486 | MH053450 |
| unigene_60653 | 5 | 0.692 | 0.598 | 0.153 | 0.137 | 0.639 | MH053451 |
| unigene_30066 | 6 | 0.588 | 0.554 | 0.064 | 0.057 | 0.498 | MH053452 |
| unigene_37667 | 3 | 0.402 | 0.320 | 0.223 | 0.205 | 0.331 | MH053453 |
| unigene_25213 | 4 | 0.578 | 0.510 | 0.135 | 0.118 | 0.515 | MH053454 |
| unigene_36790 | 13 | 0.745 | 0.582 | 0.240 | 0.219 | 0.717 | MH053455 |
| unigene_5197 | 9 | 0.638 | 0.514 | 0.219 | 0.194 | 0.611 | MH053456 |
| unigene_48018 | 5 | 0.525 | 0.506 | 0.038 | 0.035 | 0.419 | MH053457 |
| unigene_31908 | 15 | 0.700 | 0.640 | 0.080 | 0.085 | 0.659 | MH053458 |
| unigene_25861 | 4 | 0.526 | 0.523 | 0.007 | 0.006 | 0.418 | MH053459 |
| unigene_42278 | 7 | 0.745 | 0.632 | 0.165 | 0.152 | 0.704 | MH053460 |
| unigene_35652 | 10 | 0.733 | 0.597 | 0.205 | 0.186 | 0.711 | MH053461 |
| Fir_SSR25d-F | 5 | 0.549 | 0.478 | 0.126 | 0.130 | 0.475 | KF048045 |
| Fir_SSR81d-F | 3 | 0.527 | 0.398 | 0.285 | 0.244 | 0.422 | KF048051 |
| 均值 Mean | 7.278 | 0.631 | 0.546 | 0.150 | 0.135 | 0.569 |
| 群体 Population | 编号 Codes | 等位基因数 Number of allele (Na) | 期望杂合度 Expected heterozygosity (He) | 观测杂合度 Observed heterozygosity (Ho) | 等位基因 丰富度 Allelic richness (Ar) | 私有等位基因 丰富度 Private allelic richness (pAr) | 近交系数 Inbreeding coefficient (FIS) |
|---|---|---|---|---|---|---|---|
| 锦湖 Jinhu | JH | 3.778 | 0.539 | 0.719 | 3.520 | 0.320 | -0.345*** |
| 老苗圃 Laomiaopu | LMP | 3.778 | 0.543 | 0.638 | 3.540 | 0.190 | -0.189*** |
| 韶石山 Shaoshishan | SS | 3.389 | 0.544 | 0.765 | 3.370 | 0.150 | -0.409*** |
| 阳元山 Yangyuanshan | YYS | 4.444 | 0.548 | 0.673 | 3.820 | 0.220 | -0.203*** |
| 长老峰 Zhanglaofeng | ZLF | 4.389 | 0.584 | 0.665 | 3.850 | 0.380 | -0.178*** |
| 鸳鸯湖 Yuanyanghu | QWT | 4.056 | 0.533 | 0.648 | 3.580 | 0.230 | -0.198*** |
| 水库1 Shuiku1 | SK1 | 3.167 | 0.447 | 0.495 | 2.990 | 0.140 | -0.056*** |
| 水库2 Shuiku2 | SK2 | 3.778 | 0.544 | 0.712 | 3.470 | 0.130 | -0.310*** |
| 柴岭 Chailing | WT | 4.222 | 0.545 | 0.623 | 3.770 | 0.150 | -0.175*** |
| 平均值 Mean | 3.889 | 0.536 | 0.660 | 3.546 | 0.212 | -0.229 | |
Table 3 Genetic diversity parameters of the nine Firmiana danxiaensis populations
| 群体 Population | 编号 Codes | 等位基因数 Number of allele (Na) | 期望杂合度 Expected heterozygosity (He) | 观测杂合度 Observed heterozygosity (Ho) | 等位基因 丰富度 Allelic richness (Ar) | 私有等位基因 丰富度 Private allelic richness (pAr) | 近交系数 Inbreeding coefficient (FIS) |
|---|---|---|---|---|---|---|---|
| 锦湖 Jinhu | JH | 3.778 | 0.539 | 0.719 | 3.520 | 0.320 | -0.345*** |
| 老苗圃 Laomiaopu | LMP | 3.778 | 0.543 | 0.638 | 3.540 | 0.190 | -0.189*** |
| 韶石山 Shaoshishan | SS | 3.389 | 0.544 | 0.765 | 3.370 | 0.150 | -0.409*** |
| 阳元山 Yangyuanshan | YYS | 4.444 | 0.548 | 0.673 | 3.820 | 0.220 | -0.203*** |
| 长老峰 Zhanglaofeng | ZLF | 4.389 | 0.584 | 0.665 | 3.850 | 0.380 | -0.178*** |
| 鸳鸯湖 Yuanyanghu | QWT | 4.056 | 0.533 | 0.648 | 3.580 | 0.230 | -0.198*** |
| 水库1 Shuiku1 | SK1 | 3.167 | 0.447 | 0.495 | 2.990 | 0.140 | -0.056*** |
| 水库2 Shuiku2 | SK2 | 3.778 | 0.544 | 0.712 | 3.470 | 0.130 | -0.310*** |
| 柴岭 Chailing | WT | 4.222 | 0.545 | 0.623 | 3.770 | 0.150 | -0.175*** |
| 平均值 Mean | 3.889 | 0.536 | 0.660 | 3.546 | 0.212 | -0.229 | |
| 组群 Group | 期望杂合度 Expected heterozygosity (He) | 群体内遗传多样性 Genetic diversity within populations (Hs) | 等位基因丰富度 Allelic richness (Ar) | 私有等位基因 丰富度 Private allelic richness (pAr) | 标准遗传分化系数 Gene differentiation factor (GST) | 基因流 Gene flow (Nm) |
|---|---|---|---|---|---|---|
| 丹霞山 Danxiashan (DXS) | 0.552 | 0.564 | 5.371 | 1.602 | 0.075 | 6.941 |
| 南雄 Nanxiong (NX) | 0.517 | 0.527 | 4.836 | 1.065 | 0.053 | 9.326 |
Table 4 Genetic diversity parameters of the two Firmiana danxiaensis groups
| 组群 Group | 期望杂合度 Expected heterozygosity (He) | 群体内遗传多样性 Genetic diversity within populations (Hs) | 等位基因丰富度 Allelic richness (Ar) | 私有等位基因 丰富度 Private allelic richness (pAr) | 标准遗传分化系数 Gene differentiation factor (GST) | 基因流 Gene flow (Nm) |
|---|---|---|---|---|---|---|
| 丹霞山 Danxiashan (DXS) | 0.552 | 0.564 | 5.371 | 1.602 | 0.075 | 6.941 |
| 南雄 Nanxiong (NX) | 0.517 | 0.527 | 4.836 | 1.065 | 0.053 | 9.326 |
| 变异来源 Source of variation | 自由度 d.f. | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例 Percentage of variation (%) |
|---|---|---|---|---|
| 组群间 Among groups | 1 | 171.218 | 0.7352 | 15.06 |
| 群体间 Among populations | 7 | 111.824 | 0.2576 | 5.28 |
| 群体内 Within populations | 417 | 1,621.42 | 3.8883 | 79.66 |
| 总计 Total | 425 | 1,904.462 | 4.8811 | - |
Table 5 Results of analysis of molecular variance (AMOVA) from populations of Firmiana danxiaensis (P < 0.01)
| 变异来源 Source of variation | 自由度 d.f. | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例 Percentage of variation (%) |
|---|---|---|---|---|
| 组群间 Among groups | 1 | 171.218 | 0.7352 | 15.06 |
| 群体间 Among populations | 7 | 111.824 | 0.2576 | 5.28 |
| 群体内 Within populations | 417 | 1,621.42 | 3.8883 | 79.66 |
| 总计 Total | 425 | 1,904.462 | 4.8811 | - |
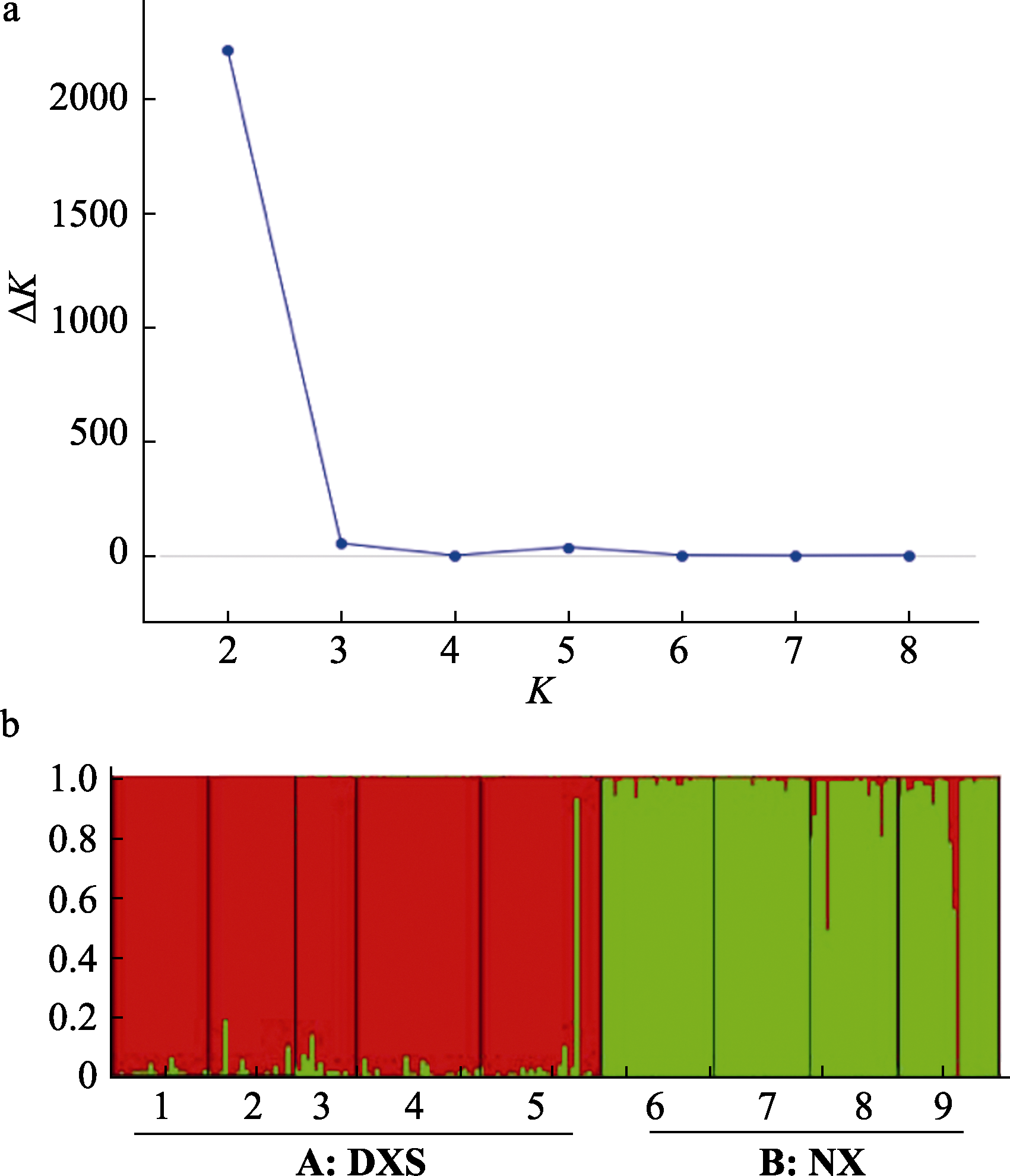
Fig. 1 Genetic structure for Firmiana danxiaensis based on EST-SSR markers. (a) The corresponding ΔK statistics calculated according to Evanno et al (2005). (b) Histogram of the structure analysis for the model with K = 2 (showing the highest ΔK). Each vertical bar represents one population. DXS represent Danxiashan group; NX represent Nanxiong group.
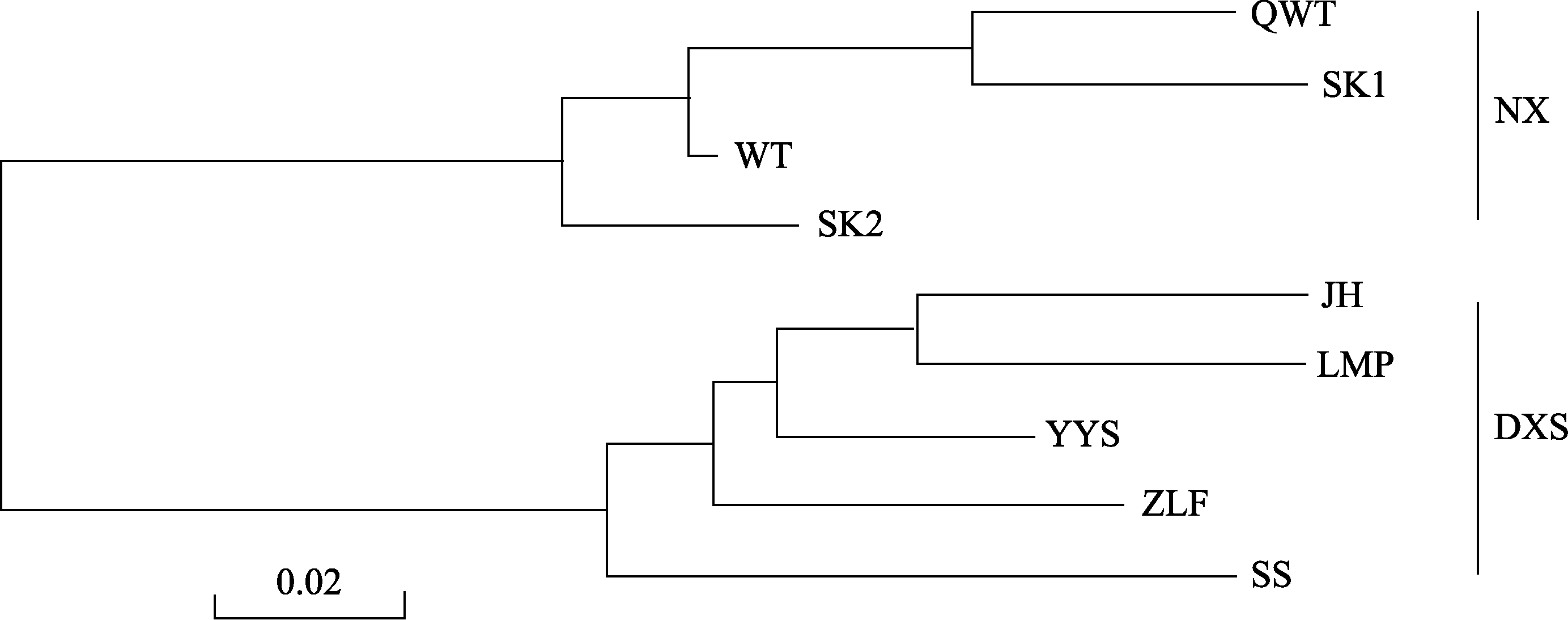
Fig. 3 Neighbor-joining network illustrating the genetic relationships among the nine Firmiana danxiaensis populations, based on Nei’s (1987) unbiased genetic distance. Meanings of abbreviations are shown in Table 1.
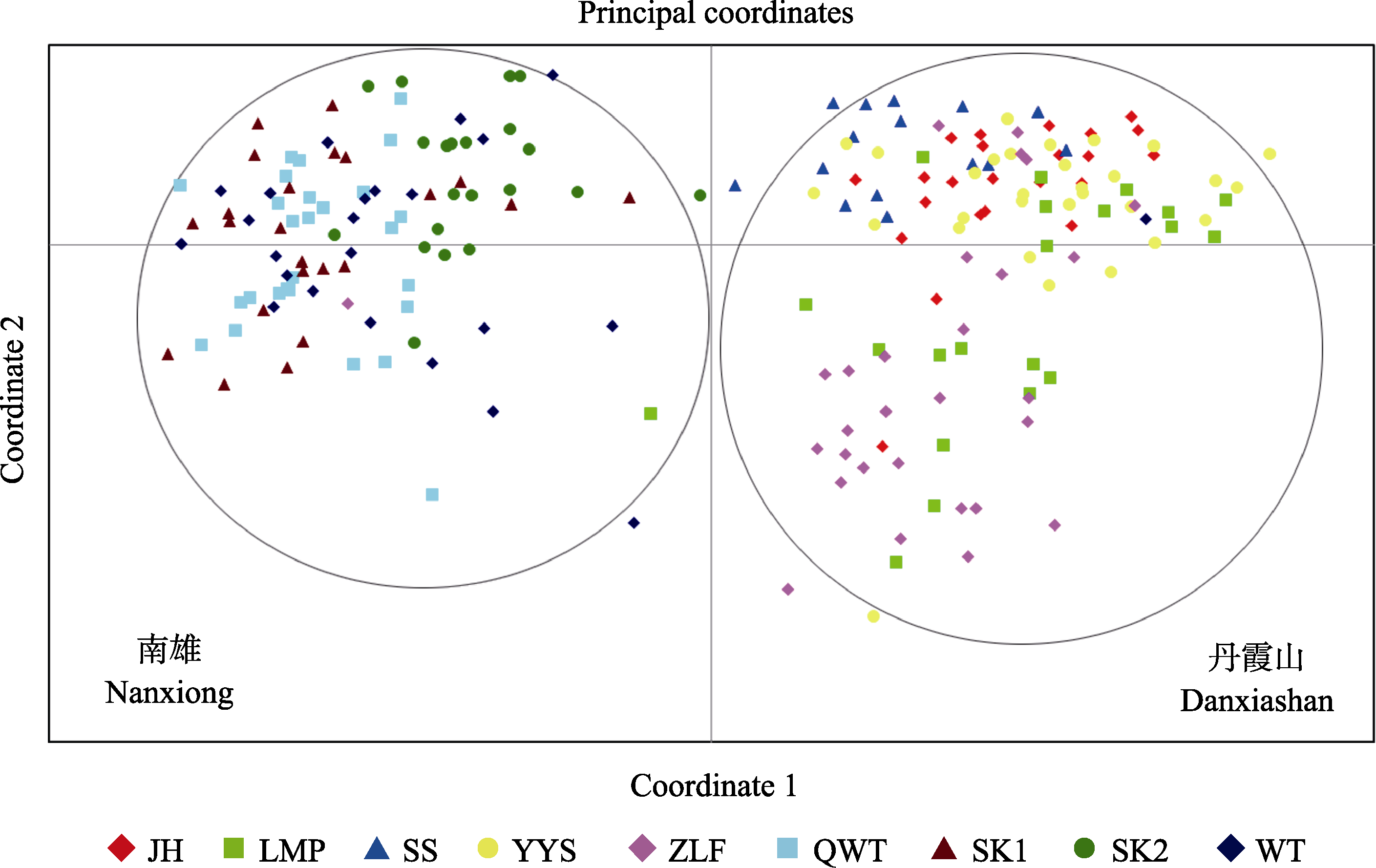
Fig. 4 Principal coordinates analysis (PCoA) of two SSR phenotypes from all nine populations of Firmiana danxiaensis. Meanings of abbreviations are shown in Table 1.
| 群体 Population | 编号 Codes | Wilcoxon检验 Wilcoxon test | |
|---|---|---|---|
| 双相突变模型 Two-phased mutation model (TPM) | 逐步突变模型 Stepwise mutation model (SMM) | ||
| 锦湖 Jinhu | JH | 0.00200 | 0.22115 |
| 老苗圃 Laomiaopu | LMP | 0.16236 | 0.95929 |
| 韶石山 Shaoshishan | SS | 0.00005 | 0.20856 |
| 阳元山 Yangyuanshan | YYS | 0.12310 | 0.95063 |
| 长老峰 Zhanglaofeng | ZLF | 0.13226 | 0.98288 |
| 鸳鸯湖 Yuanyanghu | QWT | 0.16236 | 0.94518 |
| 水库1 Shuiku1 | SK1 | 0.00237 | 0.22115 |
| 水库2 Shuiku2 | SK2 | 0.00694 | 0.63314 |
| 柴岭 Chailing | WT | 0.05935 | 0.64906 |
Table 6 Bottleneck detection for Firmiana danxiaensis
| 群体 Population | 编号 Codes | Wilcoxon检验 Wilcoxon test | |
|---|---|---|---|
| 双相突变模型 Two-phased mutation model (TPM) | 逐步突变模型 Stepwise mutation model (SMM) | ||
| 锦湖 Jinhu | JH | 0.00200 | 0.22115 |
| 老苗圃 Laomiaopu | LMP | 0.16236 | 0.95929 |
| 韶石山 Shaoshishan | SS | 0.00005 | 0.20856 |
| 阳元山 Yangyuanshan | YYS | 0.12310 | 0.95063 |
| 长老峰 Zhanglaofeng | ZLF | 0.13226 | 0.98288 |
| 鸳鸯湖 Yuanyanghu | QWT | 0.16236 | 0.94518 |
| 水库1 Shuiku1 | SK1 | 0.00237 | 0.22115 |
| 水库2 Shuiku2 | SK2 | 0.00694 | 0.63314 |
| 柴岭 Chailing | WT | 0.05935 | 0.64906 |
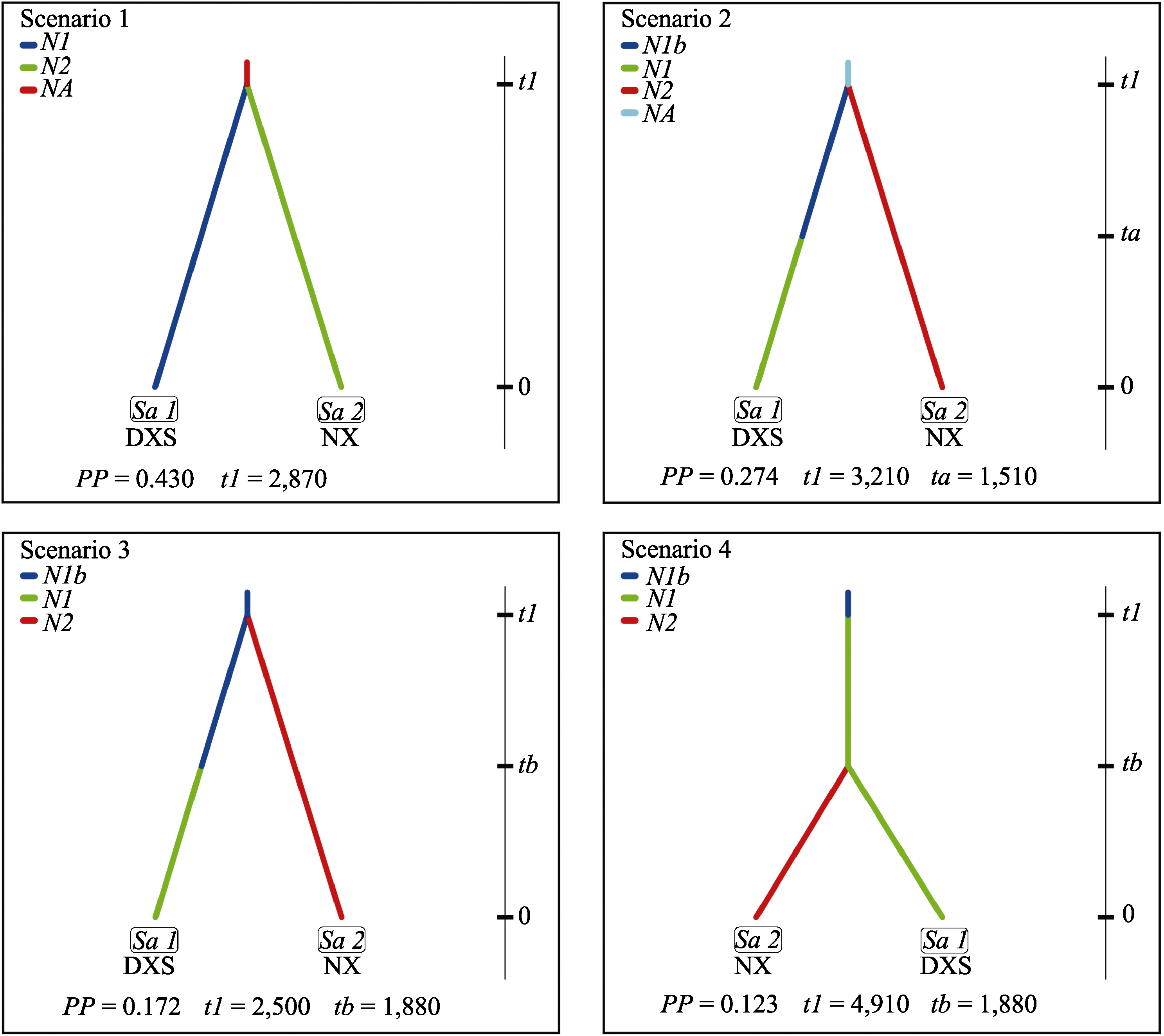
Fig. 6 Four scenarios for Firmiana danxiaensis based on Approximate Bayesian Computation (K = 2). Meanings of abbreviations are shown in Table 1. DXS represents Danxiashan group. NX represents Nanxiong group. PP represents posterior probability. N1, N2, N1b, NA represent effective population size, respectively. t1, ta, tb represent generation.
| 进化事件1 Scenario 1 | 进化事件2 Scenario 2 | 进化事件3 Scenario 3 | 进化事件4 Scenario 4 | |
|---|---|---|---|---|
| 后验概率 Posterior probability (PP) | 0.4303 | 0.2740 | 0.1724 | 0.1233 |
| 代数 Generation (t1) | 2,870 | 3,210 | 2,500 | 4,910 |
| 代数 Generation (ta) | - | 1,510 | 1,880 | - |
| 代数 Generation (tb) | - | - | - | 1,880 |
| 群体大小 Effective population size (N1b) | - | 4,830 | 6,070 | 646 |
| 群体大小 Effective population size (N1) | 7,290 | 7,800 | 8,260 | 7,220 |
| 群体大小 Effective population size (N2) | 5,550 | 5,700 | 5,430 | 5,630 |
| 群体大小 Effective population size (N3) | - | - | - | - |
| 群体大小 Effective population size (NA) | 4,450 | 4,130 | - | - |
Table 7 Median estimation of posterior distributions for each scenario based on Approximate Bayesian Computation (ABC)
| 进化事件1 Scenario 1 | 进化事件2 Scenario 2 | 进化事件3 Scenario 3 | 进化事件4 Scenario 4 | |
|---|---|---|---|---|
| 后验概率 Posterior probability (PP) | 0.4303 | 0.2740 | 0.1724 | 0.1233 |
| 代数 Generation (t1) | 2,870 | 3,210 | 2,500 | 4,910 |
| 代数 Generation (ta) | - | 1,510 | 1,880 | - |
| 代数 Generation (tb) | - | - | - | 1,880 |
| 群体大小 Effective population size (N1b) | - | 4,830 | 6,070 | 646 |
| 群体大小 Effective population size (N1) | 7,290 | 7,800 | 8,260 | 7,220 |
| 群体大小 Effective population size (N2) | 5,550 | 5,700 | 5,430 | 5,630 |
| 群体大小 Effective population size (N3) | - | - | - | - |
| 群体大小 Effective population size (NA) | 4,450 | 4,130 | - | - |
| [1] | Chen L, Zhou H, Wang MQ, Wu XT, Lin XY, Zhang Y, Wen YF (2018) Community characteristics comparison of Firmiana danxiaensis species. Chinese Wild Plant Resources, 37(2), 46-49. (in Chinese with English abstract) |
| [陈璐, 周宏, 王敏求, 武星彤, 林雪莹, 张原, 文亚峰 (2018) 丹霞梧桐群落特征比较研究. 中国野生植物资源, 37(2), 46-49.] | |
| [2] | Chen SF, Li MW, Hou RF, Liao WB, Zhou RC, Fan Q (2014) Low genetic diversity and weak population differentiation in Firmiana danxiaensis, a tree species endemic to Danxia landform in northern Guangdong, China. Biochemical Systematics & Ecology, 55, 66-72. |
| [3] | Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144, 2001-2014. |
| [4] | Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics, 30, 1187-1189. |
| [5] | Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology, 4, 129-131. |
| [6] | Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359-361. |
| [7] | Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567. |
| [8] | Frankham R, Ballou JD, Briscoe DA (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge, UK. |
| [9] | Freeland JR, Petersen SD, Kirk H (2011) Molecular Ecology, 2nd Edn. Wiley-Blackwell, West Sussex. |
| [10] | Goudet J (1995) FSTAT(Version 1.2): A computer program to calculate F-Statistics. Journal of Heredity, 86, 485-486. |
| [11] | Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society B: Biological Sciences, 351, 1291-1298. |
| [12] | Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124. |
| [13] | Harrison SP, Yu G, Takahara H, Prentice IC (2001) Palaeovegetation (communications arising): Diversity of temperate plants in East Asia. Nature, 413, 129-130. |
| [14] | Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London, 359, 183-195. |
| [15] | Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247-276. |
| [16] | Howard WR (1997) A warm future in the past. Nature, 388, 418-419. |
| [17] | Huang J, Chen ZJ, Qi DL (2015) Study on distribution of Danxia Landform in China (first). Mountain Research, 33, 385-396. (in Chinese with English abstract) |
| [黄进, 陈致均, 齐德利 (2015) 中国丹霞地貌分布(上). 山地学报, 33, 385-396.] | |
| [18] | Jakobsson M, Rosenberg NA (2007) CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801-1806. |
| [19] | Jarne P, Lagoda PJL (1996) Microsatellites, from molecules to populations and back. Trends in Ecology & Evolution, 11(10), 424-429. |
| [20] | Jing CR (1963) On the Quaternary glacial period in China. Scientific Research Papers, (1963), 253-268. (in Chinese) |
| [景才瑞 (1963) 试论中国第四纪冰期. 科学研究论文集, (1963), 253-268.] | |
| [21] | Kalinowski ST (2005) HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Resources, 5, 187-189. |
| [22] | Kou YX, Cheng SM, Tian S, Li B, Fan DM, Chen YJ, Soltis DE, Soltis PS, Zhang ZY (2016) The antiquity of Cyclocary apaliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late early Miocene. Journal of Biogeography, 43, 351-360. |
| [23] | Luo XY, Chen QH, Cai CR, Zeng HN, Ren B, Zheng WL, Feng JY (2015) Analysis on geographical elements of the community of plant species with extremely small population Firmiana danxiaensis. Journal of Shaoguan University, 36(12), 28-31. (in Chinese with English abstract) |
| [罗晓莹, 陈秋慧, 蔡纯榕, 曾浩宁, 任斌, 郑万里, 冯均尧 (2015) 极小种群植物丹霞梧桐群落的地理区系成分分析. 韶关学院学报, 36(12), 28-31.] | |
| [24] | Myers N, Mittermeier RA, Mittermeier CG, Da FG, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature, 403, 853-858. |
| [25] | Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology, 13, 1143-1155. |
| [26] | Ouyang J, Huang J (2011) Spatial distribution of Danxia landforms in China. Geospatial Information, 9(6), 55-59. (in Chinese with English abstract) |
| [欧阳杰, 黄进 (2011) 中国丹霞地貌空间分布探讨. 地理空间信息, 9(6), 55-59.] | |
| [27] | Palumbi SR (2001) Humans as the World’s Greatest Evolutionary Force. Science, 293, 1786-1790. |
| [28] | Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in Excel, population genetic software for teaching and research. Molecular Ecology Resources, 6, 288-295. |
| [29] | Peng H (2011) Danxia of China towards the world. Map, (6), 114-121. |
| (in Chinese) [彭华 (2011) 走向世界的中国丹霞. 地图, (6), 114-121.] | |
| [30] | Peng SL (2011) Comprehensive Scientific Investigation for Animal and Plant Resources in Mount Danxia, Guangdong. Science Press, Beijing. (in Chinese) |
| [彭少麟 (2011) 广东丹霞山动植物资源综合科学考察. 科学出版社, 北京.] | |
| [31] | Pritchard JK, Stephens MJ, Donnelly PJ (2000) STRUCTURE, version 2.3.3, inference of population structure using multilocus genotype data. Genetics, 155, 945-959. |
| [32] | Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Molecular Phylogenetics and Evolution, 59, 225-244. |
| [33] | Qiu YX, Guan BC, Fu CX, Comes HP (2009) Did glacials and /or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Molecular Phylogenetics and Evolution, 51, 281-293. |
| [34] | Rosenberg NA (2010) DISTRUCT: A program for the graphical display of population structure. Molecular Ecology Notes, 4, 137-138. |
| [35] | Selkoe KA, Toonen RJ (2006) Microsatellite for ecologists: A practical guide to using and evaluating microsatellite markers. Ecology Letters, 9, 615-629. |
| [36] | Takezaki N, Nei M, Tamura K (2010) POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows- interface. Molecular Biology & Evolution, 27, 747-752. |
| [37] | Tao JR (1992) Study on the fossil flowers of angiosperm. Acta Botanica Sinica, 34, 240-242.(in Chinese) |
| [陶君容 (1992) 被子植物的花化石研究. 植物学报, 34, 240-242.] | |
| [38] | Templeton AR, Robertson RJ, Brisson J, Strasburg J (2001) Disrupting evolutionary processes: The effect of the habitat fragmentation on collared lizards in the Missouri Ozarks. Proceedings of the National Academy of Sciences, USA, 98, 5426-5432. |
| [39] | Tilman D, Lehman C (2001) Human-caused environmental change: Impacts on plant diversity and evolution. Proceedings of the National Academy of Sciences, USA, 98, 5433-5440. |
| [40] | Tsumura Y, Yoshimura K, Tomaru N, Ohba K (1995) Molecular phylogeny of conifers using RFLP analysis of PCR-amplified specific chloroplast genes. Theoretical and Applied Genetics, 91, 1222-1236. |
| [41] | Wang S, Xie Y (2004) China Species Red List (Vol. 1). Higher Education Press, Beijing. (in Chinese) |
| [汪松, 解焱 (2004) 中国物种红色名录(第一卷). 高等教育出版社, 北京.] | |
| [42] | Wen YF, Han WJ, Wu S (2010) Plant genetic diversity and its influencing factors. Journal of Central South University of Forestry & Technology, 30(12), 80-87. (in Chinese with English abstract) |
| [文亚峰, 韩文军, 吴顺 (2010) 植物遗传多样性及其影响因素. 中南林业科技大学学报, 30(12), 80-87.] | |
| [43] | Xie XM, Zhang SZ, Li Y, Wu H (2003) Review of taxonomic studies on Sterculiaceae. Guihaia, 23, 311-317. (in Chinese with English abstract) |
| [解新明, 张寿洲, 李勇, 吴鸿 (2003) 梧桐科分类学研究评述. 广西植物, 23, 311-317.] | |
| [44] | Xu SJ (2002) The distribution of the Sterculiaceous plants in China. Guihaia, 22, 494-498. (in Chinese with English abstract) |
| [徐颂军 (2002) 梧桐科植物在中国的地理分布. 广西植物, 22, 494-498.] | |
| [45] | Xu SJ, Xu XH (2001) Geographical distribution of Sterculiaceae. Journal of Tropical and Subtropical Botany, 9(1), 19-30. (in Chinese with English abstract) |
| [徐颂军, 徐祥浩 (2001) 梧桐科植物的地理分布. 热带亚热带植物学报, 9, 19-30.] | |
| [46] | Xu XH, Qiu HX, Xu SJ (1987) New species and variety of Sterculiaceae from China. Journal of South China Agricultural University, 8(3), 1-5. (in Chinese with English abstract) |
| [徐祥浩, 丘华兴, 徐颂军 (1987) 中国梧桐科植物的新种和新变种. 华南农业大学学报, 8(3), 1-5.] | |
| [47] | Yao XH, Ye QG, Kang M, Huang HW (2007) Microsatellite analysis reveals interpopulation differentiation and gene flow in the endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China. New Phytologist, 176, 472-480. |
| [48] | Ye JW, Zhang Y, Wang XJ (2017) Phylogeographic history of broad-leaved forest plants in subtropical China. Acta Ecologica Sinica, 37, 5894-5904. (in Chinese with English abstract) |
| [叶俊伟, 张阳, 王晓娟 (2017) 中国亚热带地区阔叶林植物的谱系地理历史. 生态学报, 37, 5894-5904.] | |
| [49] | Ying JS (2001) Species diversity and distribution pattern of seed plants in China. Biodiversity Science, 9, 393-398. (in Chinese with English abstract) |
| [应俊生 (2001) 中国种子植物物种多样性及其分布格局. 生物多样性, 9, 393-398.] |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [11] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [12] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [13] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [14] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| [15] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()