



Biodiv Sci ›› 2021, Vol. 29 ›› Issue (12): 1607-1619. DOI: 10.17520/biods.2021273 cstr: 32101.14.biods.2021273
• Original Papers: Plant Diversity • Previous Articles Next Articles
Menghua Zhang1,2, Xianchun Zhang1,*( )
)
Received:2021-07-09
Accepted:2021-11-03
Online:2021-12-20
Published:2021-12-16
Contact:
Xianchun Zhang
Menghua Zhang, Xianchun Zhang. Species delimitation of the Selaginella delicatula group in China[J]. Biodiv Sci, 2021, 29(12): 1607-1619.
| 分子标记 Marker | 引物 Primer | 正反向 Direction | 序列 Sequence (5′-3′) | 参考文献 Reference |
|---|---|---|---|---|
| 26S | 26S 60F | 正向 Forward | TTTAAGCATATCACTAAGCGGAGG | Korall & Kenrick ( |
| 26S | 26S 380F | 正向 Forward | CCGCGAGGGAAAGATGAAAAGGAC | Korall & Kenrick ( |
| 26S | 26S 1160R | 反向 Reverse | CCAGTTCTGCTTACCAAAAATGGCCC | Korall & Kenrick ( |
| pgiC | SWPGIC-1666F | 正向 Forward | VTTYGCTTTYTGGGAYTGGG | Weststrand & Korall ( |
| pgiC | SWPGIC-2523R | 反向 Reverse | GTCGTGGTTRCTSACAATCTC | Weststrand & Korall ( |
| rbcL | rbcL-192F | 正向 Forward | CACGTGGACTACCGTTTGGA | Shalimov et al ( |
| rbcL | rbcL-1324R | 反向 Reverse | TACCCTCAAGAGCGGGATCA | Shalimov et al ( |
| psbA | psbA-169F | 正向 Forward | CCRGTAGATATTGACGGTATTC | Shalimov et al ( |
| psbA | psbA-1026R | 反向 Reverse | ATCTRGWGGGAAGTTGTGAGC | Shalimov et al ( |
| atpI | atpI-119F | 正向 Forward | CYCAGGTTCATGGACAAGTAC | Shalimov et al ( |
| atpI | atpI-540R | 反向 Reverse | GRGTATYGGGGTTGGTTG | Shalimov et al ( |
Table 1 Primers used in amplification and sequencing
| 分子标记 Marker | 引物 Primer | 正反向 Direction | 序列 Sequence (5′-3′) | 参考文献 Reference |
|---|---|---|---|---|
| 26S | 26S 60F | 正向 Forward | TTTAAGCATATCACTAAGCGGAGG | Korall & Kenrick ( |
| 26S | 26S 380F | 正向 Forward | CCGCGAGGGAAAGATGAAAAGGAC | Korall & Kenrick ( |
| 26S | 26S 1160R | 反向 Reverse | CCAGTTCTGCTTACCAAAAATGGCCC | Korall & Kenrick ( |
| pgiC | SWPGIC-1666F | 正向 Forward | VTTYGCTTTYTGGGAYTGGG | Weststrand & Korall ( |
| pgiC | SWPGIC-2523R | 反向 Reverse | GTCGTGGTTRCTSACAATCTC | Weststrand & Korall ( |
| rbcL | rbcL-192F | 正向 Forward | CACGTGGACTACCGTTTGGA | Shalimov et al ( |
| rbcL | rbcL-1324R | 反向 Reverse | TACCCTCAAGAGCGGGATCA | Shalimov et al ( |
| psbA | psbA-169F | 正向 Forward | CCRGTAGATATTGACGGTATTC | Shalimov et al ( |
| psbA | psbA-1026R | 反向 Reverse | ATCTRGWGGGAAGTTGTGAGC | Shalimov et al ( |
| atpI | atpI-119F | 正向 Forward | CYCAGGTTCATGGACAAGTAC | Shalimov et al ( |
| atpI | atpI-540R | 反向 Reverse | GRGTATYGGGGTTGGTTG | Shalimov et al ( |
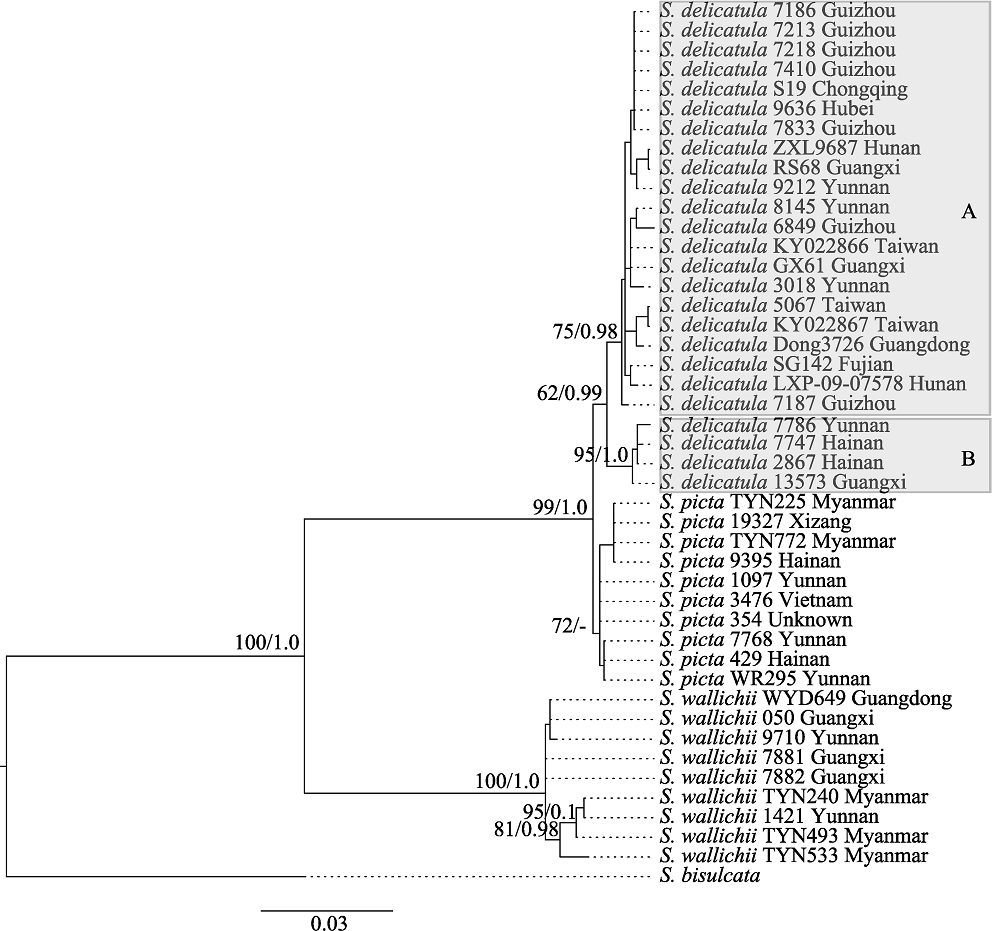
Fig. 1 Maximum likelihood tree of the Selaginella delicatula group based on the nuclear dataset (26S nrDNA + pgiC). Maximum parsimony bootstrap values and Bayesian posterior probabilities are shown on the branches (ML/PP). A dash (-) indicates no support value.
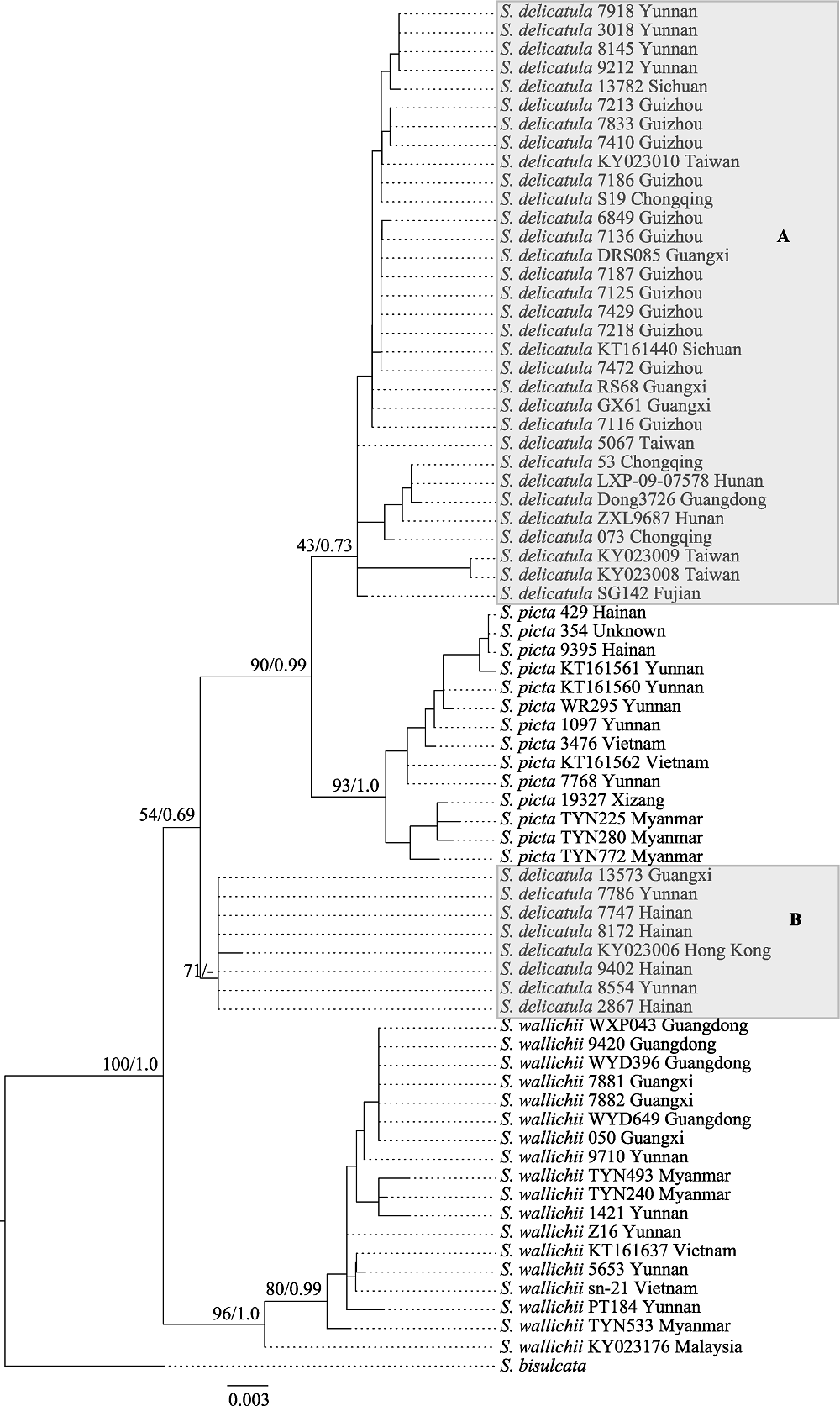
Fig. 2 Maximum likelihood tree of the Selaginella delicatula group based on the chloroplast dataset (atpI + psbA + rbcL). Maximum parsimony bootstrap values and Bayesian posterior probabilities are shown on the branches (ML/PP). A dash (-) indicates no support value.
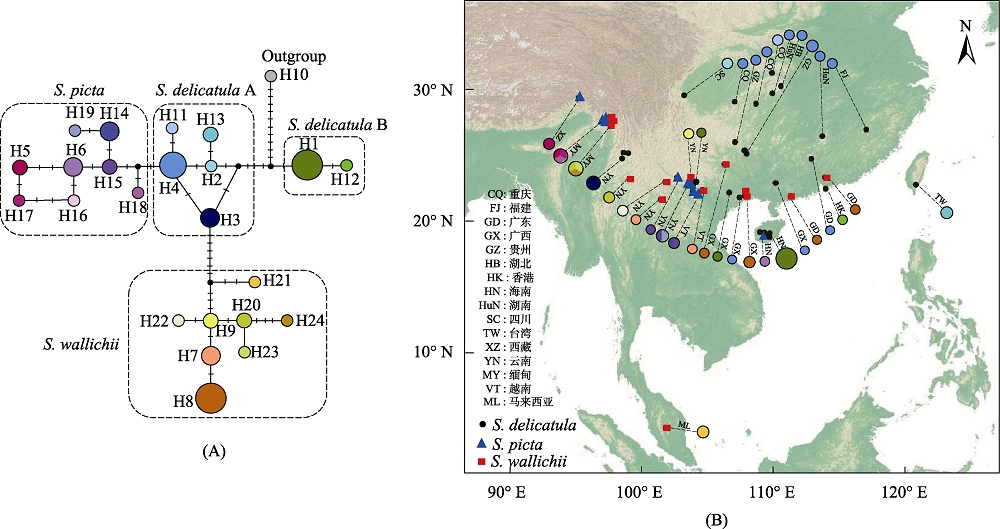
Fig. 3 Chloroplast rbcL gene haplotype network of the Selaginella delicatula group (A) and their distributions (B). CQ, Chongqing; FJ, Fujian; GD, Guangdong; GX, Guangxi; GZ, Guizhou; HB, Hubei; HK, Hong Kong; HN, Hainan; HuN, Hunan; SC, Sichuan; TW, Taiwan; XZ, Xizang; YN, Yunnan; MY, Myanmar; VT, Vietnam; ML, Malaysia.

Fig. 4 Strobili and sterile leaves of Selaginella delicatula group. (A-D) S. picata, Xizang, B. S. Li & S. Z. Cheng 02630 (PE); (E-H) S. wallichii, Yunnan, X. C. Zhang & S. Y. Dong 1421 (PE); (I-L) S. delicatula B, Hainan, S. Y. Dong 826 (PE); (M-P) S. delicatula A, Hunan, Y. H. Yan 7765 (PE). (A, E, I, M) Strobili, Scale bars = 1 mm; (B, F, J, N) Ventral leaves; (C, G, K, O) Dorsal leaves; (D, H, L, P) Axillary leaves. Scale bars = 0.2 mm.

Fig. 5 Specimens of Selaginella delicatula group. (A) S. delicatula A, Guizhou, X. C. Zhang et al. 6849 (PE); (B) S. delicatula B, Pisang Island near New Guinea, Gaudichaud 13 (Lectotype, G); (C) S. wallichii, Penang and Singapore, Dr. Wallich 128 (Isotype, E); (D) S. picta, India, Griffith sn (Syntype, K).
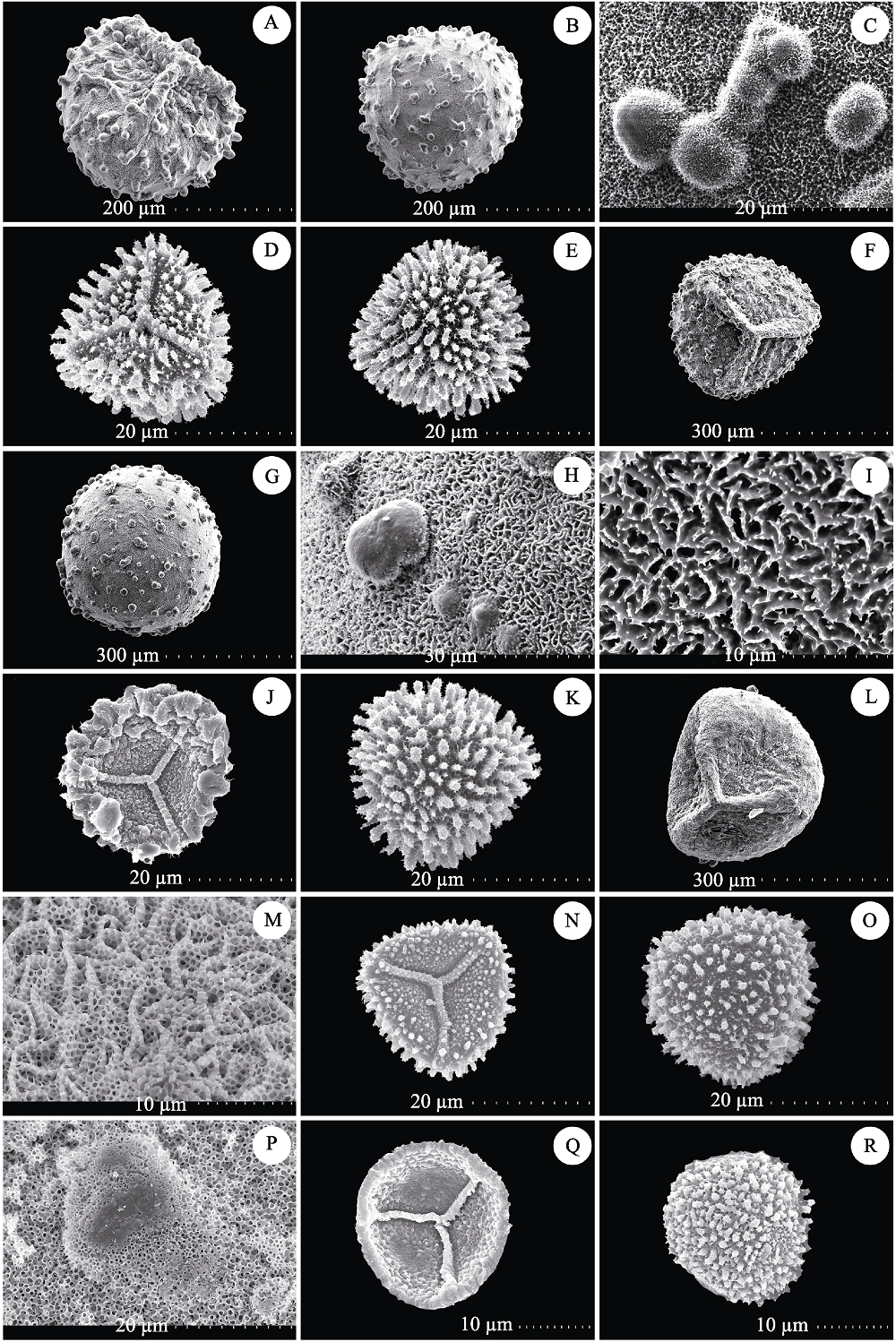
Fig. 6 Spores morphology of Selaginella delicatula group. (A-E) S. delicatula A (A-C Megaspores, D-E Microspores), Guizhou, X. C. Zhang et al. 6849 (PE); (F-K) S. delicatula B (F-I Megaspores, J-K Microspores), Hainan, X. C. Zhang 2867 (PE); (L-O) S. picta (L-M Megaspores, N-O Microspores), Yunnan, Z. Y. Li & R. Wei WR295 (PE); (P) S. wallichii (Megaspores), Guangxi, Shiwandashan Exped. 050 (PE); (Q-R) S. wallichii (Microspores), Guangxi, X. C. Zhang & Z. Y. Guo 7882 (PE).
| [1] |
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
PMID |
| [2] | Bog M, Sree KS, Fuchs J, Hoang PTN, Schubert I, Kuever J, Rabenstein A, Paolacci S, Jansen MAK, Appenroth KJ (2020) A taxonomic revision of Lemna sect. Uninerves (Lemnaceae). Taxon, 69, 56-66. |
| [3] |
Buck WR (1986) A new species of Selaginella section Articulatae (Selaginellaceae) from Paraguay. Brittonia, 38, 45-47.
DOI URL |
| [4] |
Du XY, Lu JM, Li DZ (2020) Extreme plastid RNA editing may confound phylogenetic reconstruction: A case study of Selaginella (lycophytes). Plant Diversity, 42, 356-361.
DOI URL |
| [5] |
Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792-1797.
DOI URL |
| [6] |
Gerbi SA (1986) The evolution of eukaryotic ribosomal DNA. Biosystems, 19, 247-258.
PMID |
| [7] | Jansen RK, Cai ZQ, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal JR, Kuehl JV, Boore JL (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences, USA, 104, 19369-19374. |
| [8] | Jermy AC (1986) Subgeneric names in Selaginella. Fern Gazette, 13, 117-118. |
| [9] |
Kang JS, Zhang HR, Wang YR, Liang SQ, Mao ZY, Zhang XC, Xiang QP (2020) Distinctive evolutionary pattern of organelle genomes linked to the nuclear genome in Selaginellaceae. The Plant Journal, 104, 1657-1672.
DOI URL |
| [10] |
Korall P, Kenrick P (2002) Phylogenetic relationships in Selaginellaceae based on rbcL sequences. American Journal of Botany, 89, 506-517.
DOI PMID |
| [11] |
Korall P, Kenrick P (2004) The phylogenetic history of Selaginellaceae based on DNA sequences from the plastid and nucleus: Extreme substitution rates and rate heterogeneity. Molecular Phylogenetics and Evolution, 31, 852- 864.
PMID |
| [12] |
Korall P, Kenrick P, Therrien JP (1999) Phylogeny of Selaginellaceae: Evaluation of generic/subgeneric relationships based on rbcL gene sequences. International Journal of Plant Sciences, 160, 585-594.
DOI URL |
| [13] |
Li YC, Wen J, Ren Y, Zhang JQ (2019) From seven to three: Integrative species delimitation supports major reduction in species number in Rhodiola section Trifida (Crassulaceae) on the Qinghai-Tibetan Plateau. Taxon, 68, 268-279.
DOI URL |
| [14] |
Liu BB, Campbell CS, Hong DY, Wen J (2020) Phylogenetic relationships and chloroplast capture in the Amelanchier- Malacomeles-Peraphyllum clade (Maleae, Rosaceae): Evidence from chloroplast genome and nuclear ribosomal DNA data using genome skimming. Molecular Phylogenetics and Evolution, 147, 106784.
DOI URL |
| [15] |
Ma XG, Sun WG, Zhu WD, Sun H (2017) Resolving the phylogenetic relationships and evolutionary history of the East Asian endemic genus Rodgersia (Saxifragaceae) using multilocus data. Perspectives in Plant Ecology, Evolution and Systematics, 25, 20-28.
DOI URL |
| [16] | Moore MJ, Bell CD, Soltis PS, Soltis DE (2007) Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences, USA, 104, 19363-19368. |
| [17] | Muller K, Muller J, Quandt D (2010) PhyDE-Phylogenetic Data Editor, version 0.9971. http://www.phyde.de/docu/docu.html . (accessed on 2017-06-01) |
| [18] |
Posada D (2008) jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution, 25, 1253-1256.
DOI PMID |
| [19] | R Core Team (2020) R: A Language and Environment for Statistical. R Foundation for Statistical Computing: Vienna. https://www.R-project.org/ . (accessed on 2021-04-21) |
| [20] | Rambaut A (2014) FigTree, Version 1.4.2. https://tree.bio.ed.ac.uk/software/figtree/ . (accessed on 2018-06-06) |
| [21] | Rieseberg LH, Soltis D (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants, 5, 65-84. |
| [22] |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542.
DOI PMID |
| [23] |
Rozas J, Ferrer-Mata A, Sánchez-Delbarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299-3302.
DOI URL |
| [24] |
Shalimov AP, Zhu YM, Zhang MH, Zhang XC (2019) Selaginella dianzhongensis (Selaginellaceae), a new spikemoss from China. PhytoKeys, 118, 75-87.
DOI URL |
| [25] |
Shao YZ, Chen Y, Zhang XC, Xiang QP (2020) Species delimitation and phylogeography of Abies delavayi complex: Inferred from morphological, molecular, and climatic data. Journal of Systematics and Evolution, 58, 234-246.
DOI URL |
| [26] |
Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688-2690.
DOI URL |
| [27] |
Wei R, Yang J, He LJ, Liu HM, Hu JY, Liang SQ, Wei XP, Zhao CF, Zhang XC (2021) Plastid phylogenomics provides novel insights into the infrafamilial relationship of Polypodiaceae. Cladistics, 37, 717-727.
DOI URL |
| [28] |
Weststrand S, Korall P (2016a) Phylogeny of Selaginellaceae: There is value in morphology after all!. American Journal of Botany, 103, 2136-2159.
DOI URL |
| [29] |
Weststrand S, Korall P (2016b) A subgeneric classification of Selaginella (Selaginellaceae). American Journal of Botany, 103, 2160-2169.
DOI URL |
| [30] |
Wu YD, Zhang HR, Zhang XC (2017) Selaginella guihaia (Selaginellaceae): A new spikemoss species from southern China and northern Vietnam around the Gulf of Tonkin. PhytoKeys, 80, 41-52.
DOI URL |
| [31] |
Yang J, Zhang MH, Wang YR, Yuan LX, Zhang XC (2021) An extraordinary rosette and resurrection new spikemoss, Selaginella iridescens (Selaginellaceae) from Hainan Island, China. Taxonomy, 1, 302-312.
DOI URL |
| [32] |
Ye JF, Niu YT, Feng YL, Liu B, Hai LS, Wen J, Chen ZD (2020) Taxonomy and biogeography of Diapensia (Diapensiaceae) based on chloroplast genome data. Journal of Systematics and Evolution, 58, 696-709.
DOI URL |
| [33] |
Zhang HR, Wei R, Xiang QP, Zhang XC (2020) Plastome- based phylogenomics resolves the placement of the sanguinolenta group in the spikemoss of lycophyte (Selaginellaceae). Molecular Phylogenetics and Evolution, 147, 106788.
DOI URL |
| [34] |
Zhang HR, Xiang QP, Zhang XC (2019a) The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (lycophyte). Genome Biology and Evolution, 11, 1258-1274.
DOI URL |
| [35] |
Zhang HR, Zhang XC, Xiang QP (2019b) Directed repeats co-occur with few short-dispersed repeats in plastid genome of a spikemoss, Selaginella vardei (Selaginellaceae, Lycopodiopsida). BMC Genomics, 20, 484.
DOI URL |
| [36] |
Zhang MH, Wei R, Xiang QP, Ebihara A, Zhang XC (2021) Integrative taxonomy of the Selaginella helvetica group based on morphological, molecular and ecological data. Taxon, 70, 1163-1187.
DOI URL |
| [37] | Zhang XC (2004) Selaginellaceae. In: Flora Reipublicae Popularis Sinicae, Tomus 6(3). Science Press, Beijing. (in Chinese) |
| [ 张宪春 (2004) 卷柏科. 见:中国植物志第六卷(第三分册). 科学出版社, 北京.] | |
| [38] | Zhang XC (2018) Some new records of Selaginella from China. Philippine Journal of Systematic Biology, 12, 22-23. |
| [39] | Zhang XC, Nooteboom HP, Kato M (2013) Selaginellaceae. In: Flora of China (eds Wu ZY, Raven PH, Hong DY). Science Press, Beijing & Missouri Botanical Garden Press, St. Louis. |
| [40] | Zhang XC, Shalimov AP, Kang JS, Zhang MH (2020) Selaginella subvaginata (Selaginellaceae), a new spikemoss from China. Journal of Species Research, 9, 221-232. |
| [41] |
Zhou XM, He ZR, Zhang L, Zhang LB (2015) Selaginella chuweimingii (Selaginellaceae) sp. nov. from Yunnan, China. Phytotaxa, 231, 283-288.
DOI URL |
| [42] |
Zhou XM, Zhang LB (2015) A classification of Selaginella (Selaginellaceae) based on molecular (chloroplast and nuclear), macromorphological, and spore features. Taxon, 64, 1117-1140.
DOI URL |
| [43] | Zhu WM (2006) Selaginellacea. In: Flora Yunnanica, Tomus 20 (Pterisophyta)(ed. Zhu WM). Science Press, Beijing. (in Chinese) |
| [ 朱维明 (2006) 卷柏科. 云南植物志, 第二十卷 (蕨类植物). 科学出版社, 北京.] |
| [1] | Shumei Zhang, Wei Li, Dingnan Li. Inventory of species diversity of Liaoning higher plants [J]. Biodiv Sci, 2022, 30(6): 22038-. |
| [2] | Xinyu Du, Jinmei Lu, Dezhu Li. Advances in the evolution of plastid genome structure in lycophytes and ferns [J]. Biodiv Sci, 2019, 27(11): 1172-1183. |
| [3] | Weibo Du, Yuan Lu. Diversity and distribution of lycophytes and ferns in the Loess Plateau [J]. Biodiv Sci, 2019, 27(11): 1260-1267. |
| [4] | Shiyong Dong, Zhengyu Zuo, Yuehong Yan, Jianying Xiang. Red list assessment of lycophytes and ferns in China [J]. Biodiv Sci, 2017, 25(7): 765-773. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn